SIRENA functions¶
Search functions by name at Index.
-
int addFirstRow(ReconstructInitSIRENA *reconstruct_init, fitsfile **inLibObject, double samprate, int runF0orB0val, gsl_vector *E, gsl_vector *PHEIGHT, gsl_matrix *PULSE, gsl_matrix *PULSEB0, gsl_matrix *MF, gsl_matrix *MFB0, gsl_matrix *COVAR, gsl_matrix *WEIGHT, gsl_matrix *PULSEMaxLengthFixedFilter)¶
Located in file: tasksSIRENA.cpp
This function writes the first row of the library (without intermediate AB-related values, because it would be necessary to have at least two rows=energies in the library). It also writes the FIXFILTT and FIXFILTF HDUs with the optimal filters in the time and frequency domain with fixed legnths (base-2 values) and the PRCLOFWM HDU with the precalculated values for optimal filtering and
EnergyMethod= WEIGHTM.Declare variables
Write in the first row of the library FITS file some columns with the info provided by the input GSL vectors
E,PHEIGHT,PULSE,PULSEB0,MFandMFB0(andCOVARandWEIGHTifhduPRCLOFWM= yes) (andPULSEMaxLengthFixedFilteriflargeFilter>OFLengthNotPadded, in general beingOFLengthNotPadded`=:option:`OFLength)Writing HDUs with fixed filters in time (FIXFILTT) and frequency (FIXFILTF), Tx and Fx columns respectively (calculating the optimal filters,
calculus_optimalFilter()). In time domain Tx columns are real numbers but in frequency domain Fx columns are complex numbers (so real parts are written in the first half of the column and imaginary parts in the second one)Calculate and write the pre-calculated values by using the noise weight matrix from noise intervals (M’WM)^{-1}M’W for different lengths, OFWx columns in PRCLOFWM
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
fitsfile** inLibObject
FITS object containing information of the library FITS file
double samprate
Sampling rate
int runF0orB0val
If
FilterMethod= F0 =>runF0orB0val= 1. IfFilterMethod= B0 =>runF0orB0val= 0gsl_vector* E
First energy to be included in the library
gsl_vector* PHEIGHT
Pulse height associated to the first energy to be included in the library
gsl_matrix* PULSE
Pulse template associated to the first energy to be included in the library
gsl_matrix* PULSEB0
Pulse template without baseline associated to the first energy to be included in the library
gsl_matrix* MF
Matched filter associated to the first energy to be included in the library
gsl_matrix* MFB0
Matched filter (baseline subtracted) associated to the first energy to be included in the library
gsl_matrix* COVAR
Covariance matrix associated to the first energy to be included in the library
gsl_matrix* WEIGHT
Weight matrix associated to the first energy to be included in the library
gsl_matrix* PULSEMaxLengthFixedFilter
Pulse template whose length is
largeFilterassociated to the first energy to be included in the library
-
int align(double samprate, gsl_vector **vector1, gsl_vector **vector2)¶
Located in file: tasksSIRENA.cpp
Based on [GilPita+05]
This function aligns
vector1withvector2(by delaying or moving forwardvector2) assuming thatvector1andvector2are shifted replicas of the same function.From the discrete function
![x[n] (n=0,...,N-1,N)](_images/math/949050832afa80f0e137525c0ad2fef420a0ef86.png) and according to the time shifting property of the Fourier transform:
and according to the time shifting property of the Fourier transform:![& x[n] <------> X[f]\\
& x[n-m] <------> X[f] exp(-j2\cdot\pi\cdot m/N)](_images/math/d87fbdc8e3bf353ed40f90fbcbf9cc99d967f8e2.png)
If
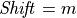 then
then 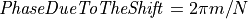 and thus,
and thus, 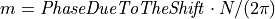
Declare variables
FFT of
vector1FFT of
vector2(Phases of the FFT_vector1 and FFT_vector2)
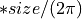
Shift between the input vectors
shiftdouble into shiftint (because we are working with samples)
Move forward or delay
vector1depending on positive or negative shift
Members/Variables
double samprate
Sampling rate
gsl_vector** vector1
GSL vector with input vector
gsl_vector** vector2
GSL with input vector which is delayed or moved forward to be aligned with
vector1
-
int calculateEnergy(gsl_vector *vector, int pulseGrade, gsl_vector *filter, gsl_vector_complex *filterFFT, int runEMethod, int indexEalpha, int indexEbeta, ReconstructInitSIRENA *reconstruct_init, int domain, double samprate, gsl_vector *Pab, gsl_matrix *PRCLWN, gsl_matrix *PRCLOFWM, double *calculatedEnergy, int numlags, double *tstartNewDev, int productSize, int tooshortPulse_NoLags)¶
Located in file: tasksSIRENA.cpp
This function calculates the energy of a pulse (
vector) depending on theEnergyMethod,OFNoise, and theFilterDomainselected from input parameters.OPTFILT and NSD (= I2R or I2RFITTED): Optimal filter = Wiener filter (see Optimal Filtering by using the noise spectral density)
Once the filter template has been created (
filterorfilterFFT), pulse height analysis is performed by aligning the template with a pulse and multiplying each point in the template by the corresponding point in the pulse. The sum of these products is the energy.In the practice, the alignment of the pulse relative to the trigger is not completely accurate, so a number of n lags could be used in order to find the peak value of the energy. The n peak values are fitted to a parabola to find the most accurate energy (
LagsOrNot) and a corrected starting time.OPTFILT and WEIGHTM (= I2R or I2RFITTED) (see Optimal Filtering by using the noise weight matrix from noise intervals)
WEIGHT and WEIGHTN (see Covariance matrices)
Members/Variables
gsl_vector* vector
Pulse whose energy has to be determined
int pulseGrade
Grade of the input pulse (to decide whether a full or only a rough estimation of energy is required).
gsl_vector* filter
Optimal filter in time domain
gsl_vector_complex* filterFFT
Optimal filter in frequency domain
int runEMethod
EnergyMethod= OPTFILT =>runEMethod= 0EnergyMethod= I2R =>runEMethod= 0EnergyMethod= I2RFITTED =>runEMethod= 0EnergyMethod= WEIGHT =>runEMethod= 1EnergyMethod= WEIGHTN =>runEMethod= 2
int indexEalpha
Index of the energy lower than the energy of the pulse which is being analyzed
int indexEbeta
Index of the energy higher than the energy of the pulse which is being analyzed
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
int domain
FilterDomain= T =>domain= 0FilterDomain= F =>domain= 1
double samprate
Sampling rate in Hz
gsl_vector* Pab
PAB column in the library
gsl_vector* PRCLWN
PCLx column in the library
gsl_vector* PRCLOFWM
OFWx column in the library
double* calculatedEnergy
Calculated energy in eV.
int numlags
Number of lags (if option:EnergyMethod = OPTFILT or I2R or I2RFITTED and
OFNoise= NSD)double tstartNewDev
Addional deviation of the starting time (if
LagsOrNot= 1)int productSize
Size of the scalar product to be calculated
int tooshortPulse_NoLags
Pulse too short to apply lags (1) or not (0)
-
int calculateIntParams(ReconstructInitSIRENA *reconstruct_init, int indexa, int indexb, double samprate, int runF0orB0val, gsl_matrix *modelsaux, gsl_matrix *covarianceaux, gsl_matrix *weightaux, gsl_vector *energycolumn, gsl_matrix **Wabaux, gsl_matrix **TVaux, gsl_vector **tEcolumn, gsl_matrix **XMaux, gsl_matrix **YVaux, gsl_matrix **ZVaux, gsl_vector **rEcolumn, gsl_matrix **Pabaux, gsl_matrix **Dabaux, gsl_matrix **PrecalWMaux, gsl_matrix **optimalfiltersabFREQaux, gsl_matrix **optimalfiltersabTIMEaux, gsl_matrix *modelsMaxLengthFixedFilteraux, gsl_matrix **PabMaxLengthFixedFilteraux)¶
Located in file: tasksSIRENA.cpp
This function calculates some intermediate scalars, vectors and matrices (WAB, TV, tE, XM, YV, ZV, rE, PAB and DAB) for the interpolation and covariance methods. See Covariance matrices reconstruction method. It is used in
readAddSortParams().Declare variables and allocate GSL vectors and matrices
Calculate intermediate scalars, vectors and matrices
Free allocated GSL vectors and matrices
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
int indexa
Lower index of the library to calculate the intermediate params (
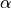 )
)int indexb
Higher index of the library to calculate the intermediate params (
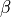 )
)double samprate
Sampling rate
int runF0orB0val
If
FilterMethod= F0 =>runF0orB0val= 1. IfFilterMethod= B0 =>runF0orB0val= 0gsl_matrix* modelsaux
GSL input matrix with model template
gsl_matrix* covarianceaux
GSL input matrix with covariance matrix
gsl_matrix* weightaux
GSL input matrix with weight matrix
gsl_vector* energycolumn
GSL input vector with list of energies
gsl_matrix** WAB
Input/output intermediate parameter
gsl_matrix** TVaux
Input/output intermediate parameter
gsl_vector** tEcolumn
Input/output intermediate parameter
gsl_matrix** XMaux
Input/output intermediate parameter
gsl_matrix** YVaux
Input/output intermediate parameter
gsl_matrix** ZVaux
Input/output intermediate parameter
gsl_vector** rEcolumn
Input/output intermediate parameter
gsl_matrix** Pabaux
Input/output intermediate parameter
gsl_matrix** Dabaux
Input/output intermediate parameter
gsl_matrix** precalWMaux
Input/output intermediate parameter
gsl_matrix** optimalfiltersabFREQaux
Input/output intermediate parameter
gsl_matrix** optimalfiltersabTIMEaux
Input/output intermediate parameter
gsl_matrix* modelsMaxLengthFixedFilteraux
Input/output intermediate parameter
gsl_matrix** PabMaxLengthFixedFilteraux
Input/output intermediate parameter
-
int calculateTemplate(ReconstructInitSIRENA *reconstruct_init, PulsesCollection *pulsesAll, PulsesCollection *pulsesInRecord, double samprate, gsl_vector **pulseaverage, double *pulseaverageHeight, gsl_matrix **covariance, gsl_matrix **weight, gsl_vector **pulseaverageMaxLengthFixedFilter)¶
Located in file: tasksSIRENA.cpp
This function calculates the template (PULSE column in the library) of non piled-up pulses. Just in case in the detection process some piled-up pulses have not been distinguished as different pulses, a pulseheights histogram is built. This function uses the pulseheights histogram (built by using the PHEIGHT column of the library), Tstart and quality to select the non piled-up pulses.
Declare and initialize variables
Before building the histogram, select the pulseheights of the pulses well separated from other pulses whose quality = 0
Create the pulseheights histogram
Calculate the pulseaverage only taking into account the valid pulses:
Check if the pulse is piled-up or not
Non piled-up pulses => Average them
Calculate covariance and weight matrices
Free allocated GSL vectors
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
PulsesCollection* pulsesAll
Collection of pulses found in the previous records
PulsesCollection* pulsesInRecord
Collection of pulses found in the current record
double samprate
Sampling rate
gsl_vector** pulseaverage
GSL vector with the pulseaverage (template) of the non piled-up pulses
double* pulseaverageHeight
Height value of the pulseaverage
gsl_matrix** covariance
GSL matrix with covariance matrix
gsl_matrix** weight
GSL matrix with weight matrix (inverse of covariance matrix)
gsl_vector** pulseaverageMaxLengthFixedFilter
GSL vector with the pulseaverage (template) whose length is
largeFilterof the non piled-up pulses
-
int calculus_optimalFilter(int TorF, int intermediate, int opmode, gsl_vector *matchedfiltergsl, long mf_size, double samprate, int runF0orB0val, gsl_vector *freqgsl, gsl_vector *csdgsl, gsl_vector **optimal_filtergsl, gsl_vector **of_f, gsl_vector **of_FFT, gsl_vector_complex **of_FFT_complex)¶
Located in file: tasksSIRENA.cpp
See description also at optimal filter chapter
This function calculates the optimal filter for a pulse whose matched filter (normalized template) is provided as input parameter,
matchedfiltergsl. An optimal filter is just a matched filter that has been adjusted based on the noise spectrum of the system.It is assumed that all pulses are scaled versions of a template. In the frequency domain (as noise can be frequency dependent), the raw data can be expressed as
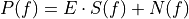 , where
, where 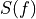 is the normalized model pulse shape in the frequency domain,
is the normalized model pulse shape in the frequency domain,
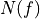 is the power spectrum of the noise and
is the power spectrum of the noise and 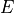 is the scalar amplitude for the photon energy.
is the scalar amplitude for the photon energy.The second assumption is that the noise is stationary, i.e., it does not vary with time. The amplitude of each pulse can then be estimated by minimizing (weighted least-squares sense) the difference between the noisy data and the model pulse shape, being the
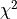 condition
to be minimized:
condition
to be minimized: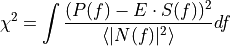
In the time domain, the amplitude is the best weighted (optimally filtered) sum of the values in the pulse
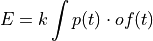
where
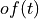 is the time domain expression of optimal filter which in frequency domain
is the time domain expression of optimal filter which in frequency domain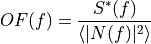
and
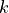 is the normalization factor to give
is the normalization factor to give 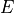 in units of energy
in units of energy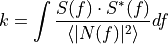
Steps:
FFT calculus of the matched filter (filter template)
Declare variables
Complex FFT values for positive and negative frequencies
FFT calculus
Generation of the frequencies (positive and negative)
Magnitude and argument for positive and negative frequencies
Free allocated GSL vectors
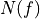
To divide
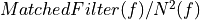 =>
=> 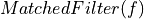 and
and 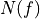 must have the same number of points
must have the same number of pointsif (
mf_size< freqgsl->size)if ((freqgsl->size)%mf_size == 0) => Decimate noise samples
else => It is necessary to work only with the positive frequencies so as not to handle the
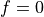 =>
=> 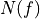 interpolation (
interpolation (interpolatePOS())
else if (
mf_size> freqgsl->size) => Error: Noise spectrum must have more samples than pulse spectrumelse if (
mf_size== freqgsl->size) => It is not necessary to do anything
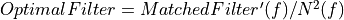
Calculus of the normalization factor
Apply the normalization factor
Inverse FFT (to get the expression of the optimal filter in time domain)
Complex
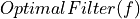 => Taking into account magnitude
=> Taking into account magnitude 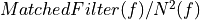 and phase given by
and phase given by 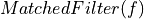
Free allocated GSL vectors
Members/Variables
int TorF
If
FilterDomain= T =>TorF= 0; IfFilterDomain= F =>TorF= 1int intermediate
If
intermediate= 0 => Do not write an intermediate file; Ifintermediate= 1 => Write an intermediate fileint opmode
gsl_vector* matchedfiltergsl
Matched filter associated to the pulse (in general, from the interpolation between two matched filters of the library)
long mf_size
Matched filter size (samples)
double samprate
Sampling rate
int runF0orB0val
If
FilterMethod= F0 =>runF0orB0val= 1. IfFilterMethod= B0 =>runF0orB0val= 0.gsl_vector* freqgsl
Frequency axis of the current noise spectral density (input)
gsl_vector* csdgsl
Current noise spectral density (input)
gsl_vector* * optimal_filtergsl
Optimal filter in time domain (output)
gsl_vector** of_f
Frequency axis of the optimal filter spectrum (output)
gsl_vector** of_FFT
Optimal filter spectrum (absolute values) (output)
gsl_vector_complex** of_FFT_complex
Optimal filter spectrum (complex values) (output)
-
int callSIRENA_Filei(char *inputFile, SixtStdKeywords *keywords, ReconstructInitSIRENA *reconstruct_init_sirena, struct Parameters par, double sampling_rate, int *trig_reclength, PulsesCollection *pulsesAll, TesEventFile *outfile)¶
Located in file: initSIRENA.c
This function calls SIRENA to build a library or reconstruct energies.
Members/Variables
char* inputFile
Input file name
SixtStdKeywords* keywords
Sixt standard keywords structure
ReconstructInitSIRENA* reconstruct_init_sirena
Parameters to run SIRENA
struct Parameters par
Input parameters
double sampling_rate
Sampling rate
int* trig_reclength
Necessary if SIRENA is going to run in THREADING mode
PulsesCollection* pulsesAll
Structure containing the detected pulses
TesEventFile* outfile
Output events FITS file
-
int callSIRENA(char *inputFile, SixtStdKeywords *keywords, ReconstructInitSIRENA *reconstruct_init_sirena, struct Parameters par, double sampling_rate, int *trig_reclength, PulsesCollection *pulsesAll, TesEventFile *outfile)¶
Located in file: initSIRENA.c
This function calls SIRENA to build a library or reconstruct energies no matter if
inputFileis only a FITS file or more (inputFile can start with ‘@’ or not).Members/Variables
char* inputFile
Input file name
SixtStdKeywords* keywords
Sixt standard keywords structure
ReconstructInitSIRENA* reconstruct_init_sirena
Parameters to run SIRENA
struct Parameters par
Input parameters
double sampling_rate
Sampling rate
int* trig_reclength
Necessary if SIRENA is going to run in THREADING mode
PulsesCollection* pulsesAll
Structure containing the detected pulses
TesEventFile* outfile
Output events FITS file
-
int checkXmls(struct Parameters *const par)¶
Located in file: initSIRENA.c
This function checks if the XML file used to build the library is the same to be used to recconstruct
Members/Variables
struct Parameters* const par
Structure containing the input parameters specified in tesreconstruction.par, teslib.par or tesrecons.par
-
int convertI2R(char *EnergyMethod, double Ibias, double Imin, double Imax, double ADU_CNV, double ADU_BIAS, double I_BIAS, double Ifit, double samprate, gsl_vector **invector)¶
Located in file: tasksSIRENA.cpp
This funcion converts the current space into a quasi-resistance space (see Quasi Resistance Space for I2R and I2RFITTED modes). The input
invectorfilled in with current values is filled in here with I2R or I2RFITTED quasi-resistances at the output.If the
ADU_CNVkeyword is in the input FITS file andinvectorcontains the ADC column data from the input FITS file: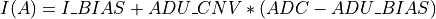 being
being 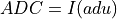 and
and ADU_CNV,ADU_BIASandI_BIASare keywords in the input FITS fileConversion according to
EnergyMethod= I2R: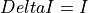
![R/R0 = [1 - (abs(DeltaI)/I\_BIAS)/(1+abs(DeltaI)/I\_BIAS)]\cdot10^5](_images/math/23e57ccecc638a557496cc94f9d5fc0effbe93f6.png)
Conversion according to
EnergyMethod= I2RFITTED: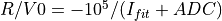 being
being 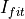 value an input parameter
value an input parameter
If the
ADU_CNVkeyword is NOT in the input FITS file andinvectorcontains the ADC column data from the input FITS file: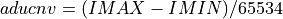 (
(IMINandIMAXare keywords in the input FITS file and 65534 the number of quantification leves)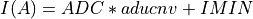 being
being 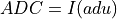
Conversion according to
EnergyMethod= I2R: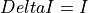
![R/R0 = [1 - (abs(DeltaI)/I0\_START)/(1+abs(DeltaI)/I0\_START)]\cdot10^5](_images/math/7af5b43b6dcb5eceda4a1cdc694eb9da8f1ef0a7.png)
Conversion according to
EnergyMethod= I2RFITTED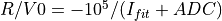 being
being 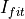 value an input parameter
value an input parameter
The
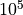 scaling factor has been included in the quasi resistance space (both I2R and I2RFITTED transformations) to avoid rounding errors when working with very small numbers.
scaling factor has been included in the quasi resistance space (both I2R and I2RFITTED transformations) to avoid rounding errors when working with very small numbers.Members/Variables
char* EnergyMethod
Quasi-resistance energy calculation method: I2R or I2RFITTED,
EnergyMethoddouble Ibias
Initial bias current (I0_START column)
double Imin
Current corresponding to 0 ADU (
IMINkeyword)double Imax
Current corresponding to maximum ADU (
IMAXkeyword)double ADU_CNV
Conversion factor (A/adu) (
ADU_CNVkeyword)double ADU_BIAS
Bias current (adu) (
ADU_BIASkeyword)double I_BIAS
Bias current (A) (
I_BIASkeyword)double samprate
Sampling rate
gsl_vector* invector
GSL vector with input signal values (ADC column of the input FITS file)
-
int createDetectFile(ReconstructInitSIRENA *reconstruct_init, double samprate, fitsfile **dtcObject, int inputPulselength)¶
Located in file: tasksSIRENA.cpp
This function creates an intermediate FITS file with some useful info (during the development phase) if the
intermediateinput parameter is set to 1.The intermediate FITS file will contain 2 HDUs:
PULSES HDU will contain some info about the found pulses: TSTART, I0 (the pulse itself), TEND, TAURISE, TAUFALL and QUALITY
TESTINFO HDU will contain columns FILDER (the low-pass filtered and differentiated records) and THRESHOLD
If file exists => Check
clobberfor overwritting. If it does not, then create it.Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
double samprate
Sampling rate
fitsfile dtcObject
Object which contains information of the intermediate FITS file (used also by
writeTestInfo()andwritePulses()).int inputPulseLength
OFLengthNotPaddedinput parameter
-
int createHisto(gsl_vector *invector, int nbins, gsl_vector **xhistogsl, gsl_vector **yhistogsl)¶
Located in file: tasksSIRENA.cpp
This function builds the histogram of the input vector.
Histogram x-axis values are the different input vector values (pulseheights)
Histogram y-axis values are the the number of cases per unit of the variable on the horizontal axis
Declare variables
It will work with the positive elements of the input vector => invectoraux2
Check if all the values of
invectorare the same => Histogram of only one binObtain invector_max and invector_min
Obtain binSize
Create histogram axis
Free allocated GSL vectors
Members/Variables
gsl_vector* invector
GSL input vector
int nbins
Number of bins to build the histogram
gsl_vector** xhistogsl
GSL vector with output histogram x-axis
gsl_vector** yhistogsl
GSL vector with output histogram y-axis
-
int createLibrary(ReconstructInitSIRENA *reconstruct_init, bool *appendToLibrary, fitsfile **inLibObject)¶
Located in file: tasksSIRENA.cpp
This function creates the pulse templates library FITS file, if it does not exist yet. Otherwise, it opens it (to add a new row).
If it exists => Open it and set appendToLibrary = true
If it does not exist => Create it and set appendToLibrary = false
Write keyword
EVENTCNT= 1 in the LIBRARY extensionWrite the whole list of input parameters in
HISTORYin the Primary extension (by usin ‘HDpar_stamp’)
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
bool appendToLibrary
Used by the function
writeLibrary()fitsfile** inLibObject
Object which contains information of the library FITS file (used also by
writeLibrary())
-
int createTPSreprFile()¶
Located in file: gennoisespec.cpp
This function creates the gennoisespec output FITS file.
Steps:
Create the noise representation file (if it does not exist already)
Create the extensions NOISE, NOISEALL and WEIGHTMS
Write keywords
-
int differentiate(gsl_vector **invector, int szVct)¶
Located in file: pulseprocess.cpp
This function applies the derivative method
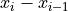 to the input vector.
to the input vector.The derivative method provides more sensitivity to handle with piled-up pulses. Moreover, little variations of the baseline will not affect.
Members/Variables
gsl_vector** invector
Input/Ouput GSL vector (non-differentiate input vector/differentiate input vector)
int szVct
Size of
invector
-
int eigenVV(gsl_matrix *matrixin, gsl_matrix **eigenvectors, gsl_vector **eigenvalues)¶
Located in file: tasksSIRENA.cpp
This funcion provides the principal eigenvectors and eigenvalues of the input matrix (at the moment, the first two eigenvalues and eigenvectors). The eigenvalues and eigenvectors are sorted in descending order and only the principal components are provided.
Calculate the eigenvectors and the eigenvalues
Sort the eigenvectors and the eigenvalues in descending order
Choose the main eigenvectors and eigenvalues (the principal components analysis). At the moment, the first two eigenvectors and eigenvalues
Members/Variables
gsl_matrix* matrixin
Input GSL matrix
gsl_matrix** eigenvectors
Subset of eigenvectors of ‘matrixin’ chosen by PCA (the first two ones)
gsl_vector** eigenvalues
Subset of eigenvalues of ‘matrixin’ chosen by PCA (the first two ones)
-
void exit_error(const char *const func, string msg, int status)¶
Located in file: genutils.cpp
This function prints out error messages and exits program.
Members/Variables
const char* const func
Function name whose error is printed
string msg
Error message to be printed
int status
Status
-
int FFT(gsl_vector *invector, gsl_vector_complex *outvector, double STD)¶
Located in file: genutils.cpp
This function calculates the FFT of the elements of a vector.
GSL library (overview of FFTs):
For physical applications it is important to remember that the index appearing in the DFT does not correspond directly to a physical frequency. If the time-step of the DFT is
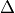 then the frequency domain includes both positive and negative frequencies, ranging from
then the frequency domain includes both positive and negative frequencies, ranging from 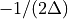 through 0 to
through 0 to 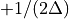 . The positive frequencies are stored from the beginning of the array up to the middle, and the negative frequencies are stored backwards from the end of the array.
. The positive frequencies are stored from the beginning of the array up to the middle, and the negative frequencies are stored backwards from the end of the array.Here is a table which shows the layout of the array data, and the correspondence between the time domain data z, and the frequency domain data x.
index
z
x = FFT(z)
0
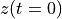
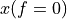
1
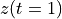
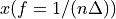
2
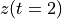
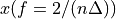
[…]
[……..]
[………………]
n/2
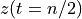
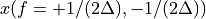
[…]
[……..]
[………………]
n-3
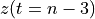
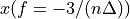
n-2
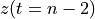
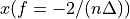
n-1
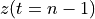
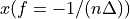
The frequency axis will be built as f = i/STD = i/(size/samprate) with i varying from 0 to size/2-1 (n=size and
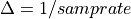 sec/sample).
sec/sample).Members/Variables
gsl_vector* invector
Input GSL vector
gsl_vector_complex* outvector
Output GSL complex vector with the FFT of
invectordouble STD
SelectedTimeDuration = (Size of
invector)/samprate
-
int FFTinverse(gsl_vector_complex *invector, gsl_vector *outvector, double STD)¶
Located in file: genutils.cpp
This function calculates the inverse FFT of the elements of a vector.
Members/Variables
gsl_vector_complex* invector
Input GSL complex vector
gsl_vector* outvector
Output GSL vector with the inverse FFT of
invectordouble STD
SelectedTimeDuration = (Size of
invector)/samprate
-
int filderLibrary(ReconstructInitSIRENA **reconstruct_init, double samprate)¶
Located in file: tasksSIRENA.cpp
This function calculates the (low-pass filter and) derivative of the models (pulse_templates) in the library (only necessary if first record), and it stores the pulse_templates_filder and the maxDERs and samp1DERs in the
reconstruct_initstructure.The maximum of the (low-pass filtered and) differentiated pulse has to be compared to the maxDERs to select the appropriate model. Or, the 1st sample out of the differentiated pulse has to be compared to the samp1DERs to select the appropriate model.
Check if it is the first record
(Low-pass filter and) differentiate the models (pulse_templates) of the library
Store the (low-pass filtered) derivatives in pulse_templates_filder
Calculate the maximum of the (low-pass filtered and) differentiated models (maxDERs)
Locate the 1st sample of the (low-pass filtered and) differentiated models (samp1DERs)
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
double samprate
Sampling rate
-
bool fileExists(const std::string &name)¶
Located in file: genutils.cpp
This function checks for file existence returning a boolean value.
Members/Variables
const std::string& name
File name
-
int fillReconstructInitSIRENAGrading(struct Parameters par, AdvDet *det, ReconstructInitSIRENA **reconstruct_init_sirena)¶
Located in file: initSIRENA.c
This function reads the grading data from the XML file and store it in themember grading of the
reconstruct_init_sirena.Members/Variables
struct Parameters par
Input parameters
AdvDet* det
Pixel detector
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
-
int filterByWavelets(ReconstructInitSIRENA *reconstruct_init, gsl_vector **invector, int length, int *onlyOnce)¶
Located in file: tasksSIRENA.cpp
This function filters the input/output signal
invector, reducing the noise level.Steps:
It is only going to work with n elements of :cpp:member:’invector’
Discrete Wavelet Transform
Sorting coefficients
Hard thresholding: n-nc coefficients are deleted (those with low energy)
Inverse DWT
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
gsl_vector** invector
Input/output signal
int length
Length of the wavelet transform
int* onlyOnce
In order to control the times to be executed
-
int findInterval(int tail_duration, gsl_vector *invector, gsl_vector *startpulse, int npin, int pulse_length, int nPF, int interval, int *ni, gsl_vector **startinterval)¶
Located in file: gennoisespec.cpp
This function finds the pulse-free intervals when the input vector has pulses. The pulse-free intervals must have a minimum length (intervalMinBins). The interval with pulse is
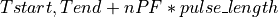 (being
(being 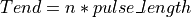 ).
).Steps:
Declare variables
- Processing if the input vector has more pulses
It looks for pulse-free intervals between pulses
- Processing if there are no more pulses in the input vector
It looks for pulse-free intervals at the end of the event and the search for more pulse-free intervals is finished
Members/Variables
int tail_duration
Length of the tail of a previous pulse
gsl_vector* invector
Input vector WITH pulses
gsl_vector* startpulse
Vector with the Tstart of all the pulses of the input vector (samples)
int npin
Number of pulses in the input vector
int pulse_length
Pulse length (samples)
int nPF
Number of pulse lengths after ending the pulse to start the pulse-free interval
int interval
Minimum length of the interval (samples)
int ni
Number of pulse-free intervals in the input vector
gsl_vector** startinterval
Vector with the starting time of each pulse-free interval (samples)
-
int findIntervalN(gsl_vector *invector, int interval, int *ni, gsl_vector **startinterval)¶
Located in file: gennoisespec.cpp
This function finds the pulse-free intervals when the input vector has NO pulses. The pulse-free intervals must have a minimum length (intervalMinBins).
Members/Variables
gsl_vector* invector
Input vector WITHOUT pulses
int interval
Minimum length of the interval (samples)
int* ni
Number of pulse-free intervals in the input vector
gsl_vector** startinterval
Vector with the starting time of each pulse-free interval (samples)
-
int findMeanSigma(gsl_vector *invector, double *mean, double *sigma)¶
Located in file: pulseprocess.cpp
This function calculates the mean and the standard deviation of the input vector.
Members/Variables
gsl_vector* invector
Input GSL vector
double* mean
Mean of the elements of
invectordouble* sigma
Standard deviation of the elements of
invector
-
int findPulsesCAL(gsl_vector *vectorin, gsl_vector *vectorinDER, gsl_vector **tstart, gsl_vector **quality, gsl_vector **pulseheight, gsl_vector **maxDERgsl, int *nPulses, double *threshold, double scalefactor, double samplingRate, int samplesup, double nsgms, double lb, double lrs, ReconstructInitSIRENA *reconstruct_init, double stopcriteriamkc, double kappamkc)¶
Located in file: pulseprocess.cpp
This function is going to find the pulses in a record (in the CALibration mode) by using the function
findTstartCAL().Steps:
Declare variables
Establish the threshold (call
medianKappaClipping())Find pulses (call
findTstartCAL())If at least a pulse is found
Get
pulseheightof each found pulse (in order to be used to build the pulse templates library)
Free allocated GSL vectors
Members/Variables
gsl_vector* vectorin
Not filtered record
gsl_vector* vectorinDER
Derivative of the (low-pass filtered)
vectoringsl_vector** tstart
Starting time of the found pulses into the record (in samples)
gsl_vector** quality
Quality of the found pulses into the record
gsl_vector** pulseheight
Pulse height of the found pulses into the record
gsl_vector** maxDERgsl
Maximum of the derivative of the found (low-pass filtered) pulses into the record
int* nPulses
Number of found pulses
double* threshold
Threshold used to find the pulses (output parameter because it is necessary out of the function)
double scalefactor
Scale factor to calculate the LPF box-car length (
scaleFactor)double samplingRate
Sampling rate
int samplesup
Number of consecutive samples over the threshold to locate a pulse (
samplesUp)double nsgms
Number of Sigmas to establish the threshold (
nSgms)double lb
Vector containing the baseline averaging length used for each pulse
double lrs
Running sum length (
LrsTin samples)ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
double stopcriteriamkc
Used in
medianKappaClipping()(%)double kappamkc
Used in
medianKappaClipping()
-
int findPulsesNoise(gsl_vector *vectorin, gsl_vector *vectorinDER, gsl_vector **tstart, gsl_vector **quality, int *nPulses, double *threshold, double scalefactor, int sizepulsebins, double samplingRate, int samplesup, double nsgms, double stopcriteriamkc, double kappamkc)¶
Located in file: gennoisespec.cpp
This function is going to find the pulses in a record by using the function
findTstartNoise()Steps:
Declare variables
Establish the threshold (call
medianKappaClipping())Find pulses (call
findTstartNoise())Free allocated GSL vectors
Members/Variables
gsl_vector* vectorin
Not filtered record
gsl_vector* vectorinDER
Derivative of the low-pass filtered
vectoringsl_vector** tstart
Starting time of the found pulses into the record (samples)
gsl_vector** quality
Quality of the found pulses into the record
int* nPulses
Number of found pulses
double* threshold
Threshold used to find the pulses (output parameter because it is necessary out of the function)
double scalefactor
Scale factor to calculate the LPF box-car length
int sizepulsebins
Size of the pulse (samples)
double samplingRate
Sampling rate
int samplesup
Number of consecutive samples over the threshold to locate a pulse
double nsgms
Number of Sigmas to establish the threshold
double stopCriteriamkc
Used in
medianKappaClipping_noiseSigma()(%)double kappamkc
Used in
medianKappaClipping_noiseSigma()
-
int FindSecondaries(int maxPulsesPerRecord, gsl_vector *adjustedDerivative, double adaptativethreshold, ReconstructInitSIRENA *reconstruct_init, int tstartFirstEvent, int *numberPulses, gsl_vector **tstartgsl, gsl_vector **flagTruncated, gsl_vector **maxDERgsl, gsl_vector **lagsgsl)¶
Located in file: pulseprocess.cpp
This function runs after
InitialTriggering()to find all the events (except the first one) in the first derivative of the (low-pass filtered) record by using the Adjusted Derivative detection method.Steps:
Declare variables
Establishing the criteria of the slope of the derivative depending on the sampling rate
It is necessary to find the tstarts…
It looks for an event and if a pulse is found, it looks for another event
It looks for an event since the beginning (or the previous event) to the end of the record. The first condition to detect an event is that the
adjustedDerivativewas over thethresholdSelect the model of the found pulse from the libary by using the 1st sample of the derivative (samp1DER)
Dot product between the detected pulse and the pulse template in 3 different lags
If maximum of the dot product found => Stop calculating dot products in more lags
If maximum of the dot product not found => Calculate dot products in more lags (number of lags is limited to 5)
If maximum of the dot product not found => tstart is the first sample crossing above the threshold (without jitter)
Average of the first 4 samples of the derivative
Find model in order to subtract
If maximum of the dot product found => Parabola analytically defined => Locate the maximum => New tstart (with jitter)
- Iterative process in order to extract the best template from the library:
samp1DER correction
Find the model from the libary by using the corrected samp1DER
Dot product in 3 lags
Locate the maximum of the parabola
samp1DER correction
Find model in order to subtract
Template correction
Average of the first 4 samples of the derivative
The second condition to detect an event is meeting the criteria of the slope of the derivative
Subtract the model from the adjusted derivative
Select the model of the found event from the libary by using the first sample of the derivative
Subtract
… Or to use the tstart provided as input parameters
Obtain the maxDERs of the events whose tstarts have been provided (by using the maximum of the derivative to find the model)
Free allocated GSL vectors
Members/Variables
int maxPulsesPerRecord
Expected maximum number of events per record in order to not allocate the GSL variables with the size of the input vector (
EventListSize)gsl_vector* adjustedDerivative
First derivative of the (low-pass filtered) record
double adaptativethreshold
Threshold
double samprate
Sampling rate
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
int tstartFirstEvent
Tstart of the first event of the record (in samples) found by
InitialTriggering()int* numberPulses
Number of found events
gsl_vector** tstartgsl
Starting time of the found events (in samples)
gsl_vector** flagTruncated
Flag indicating if the event is truncated (inside this function only initial truncated pulses are classified)
gsl_vector** maxDERgsl
Maximum of the derivative of the event
gsl_vector** samp1DERgsl
Average of the first 4 samples of the derivative of the event
gsl_vector** lagsgsl
Number of necessary lags to establish the tstart (currently limited to 5)
-
int FindSecondariesSTC(int maxPulsesPerRecord, gsl_vector *adjustedDerivative, double adaptativethreshold, ReconstructInitSIRENA *reconstruct_init, int tstartFirstEvent, int *numberPulses, gsl_vector **tstartgsl, gsl_vector **flagTruncated, gsl_vector **maxDERgsl, gsl_vector **lagsgsl)¶
Located in file: pulseprocess.cpp
This function runs after
InitialTriggering()to find all the events (except the first one) in the first derivative of the (low-pass filtered) record by using the Single Threshold Crossing method.Steps:
Declare variables
It is necessary to find the tstarts…
It looks for an event and if a pulse is found, it looks for another event
It looks for an event since the beginning (or the previous event) to the end of the record. The condition to detect an event is that the
adjustedDerivativewas over thethresholdat leastsamplesUpconsecutive samples
… Or to use the tstart provided as input parameters
Obtain the maxDERs of the events whose tstarts have been provided
Free allocated GSL vectors
Members/Variables
int maxPulsesPerRecord
Expected maximum number of events per record in order to not allocate the GSL variables with the size of the input vector (
EventListSize)gsl_vector* adjustedDerivative
First derivative of the (low-pass filtered) record
double adaptativethreshold
Threshold
double samprate
Sampling rate
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
int tstartFirstEvent
Tstart of the first event of the record (in samples) found by
InitialTriggering()int* numberPulses
Number of found events
gsl_vector** tstartgsl
Starting time of the found events (in samples)
gsl_vector** flagTruncated
Flag indicating if the event is truncated (inside this function only initial truncated pulses are classified)
gsl_vector** maxDERgsl
Maximum of the derivative of the event
gsl_vector** samp1DERgsl
Average of the first 4 samples of the derivative of the event
-
int findTstartCAL(int maxPulsesPerRecord, gsl_vector *der, double adaptativethreshold, int nSamplesUp, ReconstructInitSIRENA *reconstruct_init, int *numberPulses, gsl_vector **tstartgsl, gsl_vector **flagTruncated, gsl_vector **maxDERgsl)¶
Located in file: pulseprocess.cpp
This function scans all the values of the derivative of the (low-pass filtered) record until it finds
nSamplesUpconsecutive values (due to the noise more than 1 value is required) over the threshold. To look for more pulses, it findsnSamplesUpconsecutive values (due to the noise) under the threshold and then, it starts to scan again.Steps:
Declare variables
Allocate GSL vectors
It is possible to find the tstarts…
Obtain tstart of each pulse in the derivative:
If
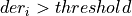 and foundPulse=false, it looks for
and foundPulse=false, it looks for nSamplesUpconsecutive samples over the thresholdIf not, it looks again for a pulse crossing over the threshold
If yes, a pulse is found (truncated if it is at the beginning)
If
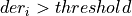 and foundPulse=true, it looks for a sample under the threshold
and foundPulse=true, it looks for a sample under the thresholdIf not, it looks again for a sample under the threshold
If yes, it looks for
nSamplesUpconsecutive samples under the threshold and again it starts to look for a pulse
… Or to use the tstart provided as input parameters
Obtain the maxDERs of the pulses whose tstarts have been provided
Members/Variables
int maxPulsesPerRecord
Expected maximum number of pulses per record in order to not allocate the GSL variables with the size of the input vector (
EventListSize)gsl_vector* der
First derivative of the (low-pass filtered) record
double adaptativethreshold
Threshold
int nSamplesUp
Number of consecutive samples over the threshold to ‘find’ a pulse (
samplesUp)ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
int* numberPulses
Number of found pulses
gsl_vector** tstartgsl
Pulses tstart (in samples)
gsl_vector** flagTruncated
Flag indicating if the pulse is truncated
gsl_vector** maxDERgsl
Maximum of the first derivative of the (low-pass filtered) record inside each found pulse
-
int findTstartNoise(int maxPulsesPerRecord, gsl_vector *der, double adaptativethreshold, int nSamplesUp, int *numberPulses, gsl_vector **tstartgsl, gsl_vector **flagTruncated, gsl_vector **maxDERgsl)¶
Located in file: gennoisespec.cpp.
This function finds the pulses tstarts in the input vector (first derivative of the filtered record).
This function scans all values the derivative of the (low-pass filtered) record until it finds nSamplesUp consecutive values (due to the noise more than 1 value is required) over the threshold. To look for more pulses, it finds nSamplesUp consecutive values (due to the noise) under the threshold and then, it starts to scan again.
Steps:
Declare variables
Allocate GSL vectors
- Obtain tstart of each pulse in the derivative:
- If
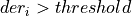 and foundPulse=false, it looks for nSamplesUp consecutive samples over the threshold
and foundPulse=false, it looks for nSamplesUp consecutive samples over the threshold If not, it looks again for a pulse crossing over the threshold
If yes, a pulse is found (truncated if it is at the beginning)
- If
- If
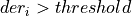 and foundPulse=true, it looks for a sample under the threshold
and foundPulse=true, it looks for a sample under the threshold If not, it looks again for a sample under the threshold
If yes, it looks for nSamplesUp consecutive samples under the threshold and again it starts to look for a pulse
- If
Members/Variables
int maxPulsesPerRecord
Expected maximum number of pulses per record in order to not allocate the GSL variables with the size of the input vector
gsl_vector* der
First derivative of the (low-pass filtered) record
double adaptativethreshold
Threshold
int nSamplesUp
Number of consecutive samples over the threshold to ‘find’ a pulse
int* numberPulses
Number of found pulses
gsl_vector** tstartgsl
Pulses tstart (samples)
gsl_vector** flagTruncated
Flag indicating if the pulse is truncated (inside this function only initial truncated pulses are classified)
gsl_vector** maxDERgsl
Maximum of the first derivative of the (low-pass filtered) record inside each found pulse
-
int find_Esboundary(double maxDER, gsl_vector *maxDERs, ReconstructInitSIRENA *reconstruct_init, int *indexEalpha, int *indexEbeta, double *Ealpha, double *Ebeta, double margin)¶
Located in file: tasksSIRENA.cpp.
This function provides the indexes of the two energies which straddle the pulse energy, by comparing the maximum value of the pulse derivative (
maxDER) to the list of maximums in the library (maxDERs).It finds the two embracing
maxDERsin the calibration library:If
maxDERis lower than the lowestmaxDERsin the library =>indexEalpha=indexEbeta= 0If
maxDERis higher than the highestmaxDERsin the library =>indexEalpha=indexEbeta= Number of templates-1
Members/Variables
double maxDER
Max value of the derivative of the (filtered) pulse whose embracing energies are being sought
gsl_vector* maxDERs
GSL vector with the maximum values of the derivatives of the templates in the library to be compared with the pulse being analysed
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values). In particular, this function uses the info in the library about the energies
int* indexEalpha
Index of the energy lower than the energy of the pulse which is being analyzed
int* indexEbeta
Index of the energy higher than the energy of the pulse which is being analyzed
double* Ealpha
Energy (in eV) which straddle the
maxDERin the lower limitdouble* Ebeta
Energy (in eV) which straddle the
maxDERin the higher limitdouble margin
Margin to be applied when several energies in the library to choose the proper filter (hardcoded in LibraryCollection in integraSIRENA.cpp)
-
int find_matchedfilterDAB(int runF0orB0val, double maxDER, gsl_vector *maxDERs, int preBuffer, ReconstructInitSIRENA *reconstruct_init, gsl_vector **matchedfilterFound, gsl_vector **PabFound, double *Ealpha, double *Ebeta, double margin)¶
Located in file: tasksSIRENA.cpp
This function selects the proper matched filter (normalized template) from the calibration library from column DAB (or from column MF if only one energy included in the library) by comparing the maximum value of the pulse derivative (
maxDER) to the list of maximums in the library (maxDERs) for the DAB interpolation method (see optimal filter chapter). It also selects the proper row from the column PAB.It finds the two embracing
maxDERsin the calibration library:Members/Variables
int runF0orB0val
If
FilterMethod= F0 =>runF0orB0val= 1. IfFilterMethod= B0 =>runF0orB0val= 0double maxDER
Max value of the derivative of the (filtered) pulse whose matched filter is being sought
gsl_vector* maxDERs
GSL vector with the maximum values of the derivatives of the templates in the library to be compared with the pulse being analysed
int preBuffer
preBuffer to work with in the particular pulse
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
gsl_vector** matchedfilterFound
GSL vector with the matched filter selected
gsl_vector** PabFound
PAB column from the library
double* Ealpha
Energy (in eV) which straddle the
maxDERin the lower limitdouble* Ebeta
Energy (in eV) which straddle the
maxDERin the higher limitdouble margin
Margin to be applied when several energies in the library to choose the proper filter (hardcoded in LibraryCollection in integraSIRENA.cpp)
-
int find_model_energies(double energy, ReconstructInitSIRENA *reconstruct_init, gsl_vector **modelFound)¶
Located in file: pulseprocess.cpp
This function uses
energyin order to choose the proper pulse template (pulse_templates_B0) of the calibration library.In general, it finds the two energies wich straddle
energyin the calibration library and interpolates (interpolate_model()):Members/Variables
double energy
Energy of the pulse whose pulse template is being sought
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values). In particular, this function uses the energies of the models (energies) and their templates (pulse_templates), the number of templates in the library (ntemplates), the template duration (template_duration) and the pulse_templates_B0.
gsl_vector** modelFound
Found template of the pulse whose energy is
energy
-
int find_model_maxDERs(double maxDER, ReconstructInitSIRENA *reconstruct_init, gsl_vector **modelFound)¶
Located in file: pulseprocess.cpp
This function uses the maximum of the derivative of the (filtered) pulse (
maxDER) in order to choose the proper pulse template (pulse_templates_filder) of the calibration library.In general, it finds the two maxDER which straddle
maxDERin the calibration library and interpolates (interpolate_model()):Members/Variables
double maxDER
Maximum of the derivative of the (filtered) pulse whose pulse template is being sought
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values). In particular, this function uses the number of templates in the library (ntemplates), the template duration (template_duration), the filtered and differentiated templates (pulse_templates_filder) and the maxDERs of the templates
gsl_vector** modelFound
Found template of the pulse whose maximum of the derivative of the filtered version is
maxDER
-
int find_model_samp1DERs(double samp1DER, ReconstructInitSIRENA *reconstruct_init, gsl_vector **modelFound)¶
Located in file: pulseprocess.cpp
This function uses the 1st sample of the derivative of the filtered pulse (
samp1DER) in order to choose the proper pulse template (pulse_templates_filder) of the calibration library.It finds the two
samp1DERcloser in the calibration library and interpolates (interpolate_model())Members/Variables
double samp1DER
1st sample of the derivative of the filtered pulse whose pulse template is being sought
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values). In particular, this function uses the 1st samples of the derivative of the models (samp1DERs) and their derived templates (pulse_templates_filder), the number of templates in the library (ntemplates) and the template duration (template_duration).
gsl_vector** modelFound
Found template of the pulse whose 1st sample of the derivative of the filtered pulse is
samp1DER
-
int find_optimalfilterDAB(double maxDER, gsl_vector *maxDERs, ReconstructInitSIRENA *reconstruct_init, gsl_vector **optimalfilterFound, gsl_vector **PabFound, double *Ealpha, double *Ebeta, double margin)¶
Located in file: tasksSIRENA.cpp
(or ‘Tx’or ‘Fx’columns if only one energy included in * the library) This function selects the proper optimal filter from the calibration library columns ABTx or ABFx (or from Tx or Fx**columns if only one energy included in the library) by comparing the maximum value of the pulse derivative (:cpp:member:`maxDER`) to the list of maximums in the library (:cpp:member:`maxDERs`). It also selects the proper row from the column **PAB.
It finds the two embracing
maxDERsin the calibration library:Members/Variables
double maxDER
Max value of the derivative of the (filtered) pulse whose optimal filter is being sought
gsl_vector* maxDERs
GSL vector with the maximum values of the derivatives of the templates in the library to be compared with the pulse being analysed
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values). In particular, this function uses the info in the library (optimal_filters)
gsl_vector** optimalfilterFound
GSL vector with the optimal filter selected
gsl_vector** PabFound
PAB column from the library
double* Ealpha
Energy (in eV) which straddle the
maxDERin the lower limitdouble* Ebeta
Energy (in eV) which straddle the
maxDERin the higher limitdouble margin
Margin to be applied when several energies in the library to choose the proper filter (hardcoded in LibraryCollection in integraSIRENA.cpp)
-
int find_prclofwm(double maxDER, gsl_vector *maxDERs, ReconstructInitSIRENA *reconstruct_init, gsl_vector **PRCLOFWMFound, double *Ealpha, double *Ebeta, double margin)¶
Located in file: tasksSIRENA.cpp
When
EnergyMethod= OPTFILT andOFNoise= WEIGHTM this function selects the proper precalculated values (OFWx) from the calibration PRCLOFWM HDU of the library by comparing the maximum value of the pulse derivative (maxDER) to the list of maximums in the library (maxDERs) for reconstruct_init->OFLib = 1.It finds the two embracing
maxDERsin the calibration library:Members/Variables
double maxDER
Max value of the derivative of the (filtered) pulse whose optimal filter is being sought
gsl_vector* maxDERs
GSL vector with the maximum values of the derivatives of the templates in the library to be compared with the pulse being analysed
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
gsl_vector** PRCLOFWMFound
GSL vector with some precalculated selected
double* Ealpha
Energy (in eV) which straddle the
maxDERin the lower limitdouble* Ebeta
Energy (in eV) which straddle the
maxDERin the higher limitdouble margin
Margin to be applied when several energies in the library to choose the proper filter (hardcoded in LibraryCollection in integraSIRENA.cpp)
-
int find_prclwn(double maxDER, gsl_vector *maxDERs, ReconstructInitSIRENA *reconstruct_init, gsl_vector **PRCLWNFound, gsl_vector **PabFound, double *Ealpha, double *Ebeta, double margin)¶
Located in file: tasksSIRENA.cpp
When
EnergyMethod= WEIGHTN this function selects the proper precalculated values (PCLx) from the PRECALWN HDU of the calibration library by comparing the maximum value of the pulse derivative (maxDER) to the list of maximums in the library (maxDERs) for the reconstruct_init->OFLib = 1. It also selects the proper row from the column PAB.It finds the two embracing
maxDERsin the calibration library:Members/Variables
double maxDER
Max value of the derivative of the (filtered) pulse whose optimal filter is being sought
gsl_vector* maxDERs
GSL vector with the maximum values of the derivatives of the templates in the library to be compared with the pulse being analysed
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
gsl_vector** PRCLWNFound
GSL vector with the precalculated values selected
gsl_vector** PabFound
PAB column from the library
double* Ealpha
Energy (in eV) which straddle the
maxDERin the lower limitdouble* Ebeta
Energy (in eV) which straddle the
maxDERin the higher limitdouble margin
Margin to be applied when several energies in the library to choose the proper filter (hardcoded in LibraryCollection in integraSIRENA.cpp)
-
extern_C_void freeOptimalFilterSIRENA(OptimalFilterSIRENA *OFilterColl)¶
Located in file: integraSIRENA.cpp
Destructor of OptimalFilterSIRENA structure.
Members/Variables
OptimalFilterSIRENA* OFilterColl
Instance of OptimalFilterSIRENA structure
-
extern_C_void freeReconstructInitSIRENA(ReconstructInitSIRENA *reconstruct_init)¶
Located in file: integraSIRENA.cpp
Destructor of ReconstructInitSIRENA structure.
Members/Variables
ReconstructInitSIRENA* reconstruct_init
Instance of ReconstructInitSIRENA structure
-
extern_C_void freePulsesCollection(PulsesCollection *PulsesColl)¶
Located in file: integraSIRENA.cpp
Destructor of PulsesCollection structure.
Members/Variables
PulsesCollection* PulsesColl
Instance of PulsesCollection structure
-
int fromGslMatrix(void **buffer, gsl_matrix **matrix, int type)¶
Located in file: inoututils.cpp
The function puts the values of the input GSL matrix into an output buffer.
Members/Variables
void** buffer
Output buffer
gsl_matrix** matrix
Input GSL matrix
int type
FITS type (TINT, TSHORT, TDOUBLE, etc.)
-
int fromGslVector(void **buffer, gsl_vector **array, int type)¶
Located in file: inoututils.cpp
The function puts the values of the input GSL vector into an output buffer.
Members/Variables
void** buffer
Output buffer
gsl_vector** array
Input GSL vector
int type
FITS type (TINT, TSHORT, TDOUBLE, etc.)
-
int gennoisespec_main()¶
Located in file: gennoisespec.cpp
This function calculates the current noise spectral density. If there are pulses in a record, the pulses are rejected and it is going to look for pulse-free intervals of a given size (intervalMinBins). If there are no pulses in a record, the event is divided into pulse-free intervals of a given size (intervalMinBins). It is going to look for pulse-free intervals, calculate their FFT(not filtered data) and average them.
Another facillity is calculate the weight matrix of the noise (in fact, weight matrixes of the noise of different lengths).
- The output FITS file (_noisespec) contains three columns in two extensions, NOISE and NOISEALL:
FREQ: Frequency
CSD: Current noise spectral density: Amount of current per unit (density) of frequency (spectral), as a function of the frequency
SIGMACSD: Standard error of the mean (filled out with 0’s at the moment)
There is also other extension, WEIGHTMS, where the weight matrices of the noise are stored.
Steps:
Reading all programm parameters by using PIL
Open input FITS file
Check if input FITS file have been simulated with TESSIM or XIFUSIM
To calculate aducnv (conversion factor between arbitrary units and A)…
…or read
ADU_CNV,I_BIASandADU_BIASGet structure of input FITS file columns
Read info to transform to resistance space
Read and check other input keywords
Read other necessary keywords from ANY HDU
- Calculate the sampling rate
By using keywords in input FITS file (from
DELTATorTCLOCK``+``DEC_FACorNUMROW``+``P_ROW)If necessary read the sampling rate from input FITS file (from the
HISTORYin the Primary HDU)If not possible, provide an error message to include DELTAT (inverse of sampling rate) in the input FITS file
Initialize variables and transform from seconds to samples
Declare variables
Create structure to run Iteration
- Read columns (TIME and ADC)
Called iteration function:
inDataIterator()
Close input FITS file
- Generate CSD representation
Applying
medianKappaClipping_noiseSigma()in order to remove the noise intervals with a high sigma (ifrmNoiseInterval= yes)FFT calculus (EventSamplesFFT)
Add to mean FFT samples
Current noise spectral density
Extra normalization (further than the FFT normalization factor,1/n) in order to get the apropriate noise level provided by Peille (54 pA/rHz)
Load in noiseIntervals only those intervals with a proper sigma (if
rmNoiseInterval= yes) and NumMeanSamples = cnt (in order not to change excesively the code when weightMS)Generate WEIGHT representation
Create output FITS File: GENNOISESPEC representation file
Write extensions NOISE, NOISEALL and WEIGHTMS (call
writeTPSreprExten())Free allocated GSL vectors
Close output FITS file
Free memory
Finalize the task
The parameters (struct Parameters par) read by
getpar_noiseSpec()are:char inFile
Name of the input FITS file
char outFile
Name of the output FITS file
int intervalMinSamples
Length of a pulse-free interval to use (samples) = intervalMinBins
int nplPF
Number of pulse lengths after ending the pulse (Tend) to start the pulse-free interval
int nintervals
Number of pulse-free intervals to use to calculate the Noise Spectral Density
double scaleFactor
Scale factor to apply in order to calculate the LPF box-car length
int samplesUp
Consecutive samples over the threshold to locate a pulse
double nSgms
Number of Sigmas to establish the threshold
int pulse_length
Pulse length (samples)
double LrsT
Running sum length (seconds)
double LbT
Baseline averaging length (seconds)
char weightMS
Calculate and write the weight matrixes if weightMS=yes
char EnergyMethod
Transform to resistance space (I2R, I2RFITTED) or not (OPTFILT)
double Ifit
Constant to apply the I2RFITTED conversion (adu)
char clobber
Re-write output files if clobber=yes
int matrixSize
Size of noise matrix if only one to be created
char rmNoiseIntervals
Remove some noise intervals before calculating the noise spectrum if rmNoiseIntervals=yes
-
int getB(gsl_vector *vectorin, gsl_vector *tstart, int nPulses, gsl_vector **lb, int sizepulse, gsl_vector **B, gsl_vector **rmsB)¶
Located in file: pulseprocess.cpp
This function calculates the sum,
B, oflbdigitized data samples of a pulse-free interval immediately before each pulse. If the pulse-free interval before the current pulse is lower thanlb,Bis calculated with the available number of samples. If there is not a pulse-free interval before the pulse, it is looked for it after the current pulse. The number of samples of the pulse-free interval used to calculateBis stored in thelbvector.Steps:
First of all, the auxiliary variable Baux is initialized to -999 and all the elements of the
lbvector are equal to theLbTinput parameter in samples. Then, the code is divided into 2 if statements:When the current pulse is the first pulse into the record:
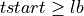 => Sum lb samples
=> Sum lb samples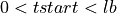 => Sum the available number of samples (although the available number of samples was lower than lb)
=> Sum the available number of samples (although the available number of samples was lower than lb)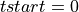 => If there is not a pulse-free interval before the pulse, it is looked for it after the current pulse
=> If there is not a pulse-free interval before the pulse, it is looked for it after the current pulse
When the current pulse is not the first pulse into the record:
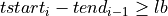 => Sum lb samples
=> Sum lb samples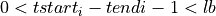 => Sum the available number of samples (although the available number of samples was lower than lb)
=> Sum the available number of samples (although the available number of samples was lower than lb)If there is not a pulse-free interval before the pulse, it is looked for it after the current pulse
If Baux is still -999, a pulse-free interval can not be found to apply the running sum filter. This has to be taken into account, out of the function, to try to get a usable
B.Members/Variables
gsl_vector* vectorin
Input record
gsl_vector* tstart
Starting time of the pulses into the record
int nPulses
Number of pulses into the record
gsl_vector** lb
Vector containing the baseline averaging length used for each pulse
int sizepulse
Size of the pulse in samples
gsl_vector** B
In general, sum of the Lb digitized data samples (
LbTinput parameters in samples) of a pulse-free interval immediately before the current pulsegsl_vector** rmsB
In general, rms of the baseline related to a pulse-free interval immediately before the current pulse
-
LibraryCollection *getLibraryCollection(const char *const filename, int opmode, int hduPRECALWN, int hduPRCLOFWM, int largeFilter, char *filter_domain, int pulse_length, char *energy_method, char *ofnoise, char *filter_method, char oflib, char **ofinterp, double filtEev, int lagsornot, int preBuffer, gsl_vector *pBi, gsl_vector *posti, int *const status)¶
Located in file: integraSIRENA.cpp
This function creates and retrieves a LibraryCollection from a file.
Create LibraryCollection structure
Open FITS file in READONLY mode (move to the first HDU) and get number of templates (rows)
Allocate library structure
Get PULSE and MF column numbers (depending the different options)
Get template duration
Allocate library structure (cont.)
Get matched filter duration
Read different columns and populate the LibraryCollection structure
Added new code to handle the new HDUs FIXFILTF, FIXFILTT, PRECALWN and PRCLOFWM
Free allocated GSL vectors and matrices
Members/Variables
const char* const filename
File with library information
int opmode
Calibration run (0) or energy reconstruction run (1),
opmodeint hduPRECALWN
Add or not the PRECALWN HDU in the library file (yes/no) (only for library creation,
opmode= 0),hduPRECALWNint hduPRCLOFWM
Add or not the PRCLOFWM HDU in the library file (yes/no) (only for library creation,
opmode= 0),hduPRCLOFWMint largeFilter
Length of the longest fixed filters (only for library creation,
opmode= 0),largeFilterchar* filter_domain
Filtering Domain: Time (T) or Frequency (F),
FilterDomainint pulse_length
Pulse length,
OFLengthNotPaddedchar* energy_method
Energy calculation Method: OPTFILT, WEIGHT, WEIGHTN, I2R, I2RFITTED or PCA,
EnergyMethodchar* ofnoise
For optimal filtering : NSD or WEIGHTM,
OFNoisechar* filter_method
Filtering Method: F0 (deleting the zero frequency bin) or B0 (deleting the baseline),
FilterMethodchar oflib
Work or not with a library with optimal filters (1/0)
char** ofinterp
Optimal Filter by using the Matched Filter or the DAB as matched filter (MF/DAB) It has been fixed in tesreconstruction as DAB (but it would be possible to work with MF)
double filtEev
Energy of the filters of the library to be used to calculate energy (only for OPTFILT, I2R and I2RFITTED),
filtEeVint lagsornot
Lags (1) or no lags (0)
int preBuffer
Using preBuffer (1) or not using preBuffer (0)
gsl_vector pBi
Vector with the preBuffer values read from the XML file
gsl_vector posti
Vector with the post values read from the XML file
int* const status
Input/output status
-
NoiseSpec *getNoiseSpec(const char *const filename, int opmode, int hduPRCLOFWM, char *energy_method, char *ofnoise, char *filter_method, int *const status)¶
Located in file: integraSIRENA.cpp
This function creates and retrieves a NoiseSpec from a file.
Create NoiseSpec structure
Open FITS file, move to the NOISE, NOISEALL and WEIGHTMS HDUs and get necessary keywords
Allocate NoiseSpec structure
Get noise spectrum (CSD), and noise frequencies (FREQ) column numbers
Read column CSD and save it into the structure
Read column FREQ and save it into the structure
Read columns Wx with the noise weight matrix from noise intervals and save them into the structure
Return noise spectrum
Members/Variables
const char* const filename
File name with noise
int opmode
Calibration run (0) or energy reconstruction run (1),
opmodeint hduPRCLOFWM
Add or not the PRCLOFWM HDU in the library file (yes/no) (only for library creation,
opmode= 0),hduPRCLOFWMchar* energy_method
Energy calculation Method: OPTFILT, WEIGHT, WEIGHTN, I2R, I2RFITTED or PCA,
EnergyMethodchar* ofnoise
For optimal filtering: NSD or WEIGHTM,
OFNoisechar* filter_method
Filtering Method: F0 (deleting the zero frequency bin) or B0 (deleting the baseline),
FilterMethodint* const status
Input/Output status
-
int getpar(struct Parameters *const par)¶
Located in file: tesreconstruction.c
This function gets the input parameter from the command line or their default values from the tesreconstruction.par file
Members/Variables
struct Parameters* const par
Structure containing the input parameters specified in tesreconstruction.par
-
int getpar_noiseSpec(struct Parameters *const par)¶
Located in file: gennoisespec.cpp
This function gets the input parameter from the command line or their default values from the gennoisespec.par file
Members/Variables
struct Parameters* const par
Structure containing the input parameters specified in gennoisespec.par
-
int getpar_teslib(struct Parameters *const par)¶
Located in file: teslib.c
This function gets the input parameter from the command line or their default values from the teslib.par file
Members/Variables
struct Parameters* const par
Structure containing the input parameters specified in teslib.par
-
int getpar_tesrecons(struct Parameters *const par)¶
Located in file: tesrecons.c
This function gets the input parameter from the command line or their default values from the tesrecons.par file
Members/Variables
struct Parameters* const par
Structure containing the input parameters specified in tesrecons.par
-
int getPulseHeight(gsl_vector *vectorin, double tstart, double tstartnext, int lastPulse, double lrs, double lb, double B, int sizepulse, double *pulseheight)¶
Located in file: pulseprocess.cpp
This function estimates the pulse height of a pulse by using a running sum filter. It extracts from the record,
vectorin, the pulse whose pulse height is going to be estimated by usingRS_filter().Steps:
Declare variables
Extracting from the record the pulse whose pulse height is going to be estimated
Apply the running sum filter
Members/Variables
gsl_vector* vectorin
Not filtered record
double tstart
Starting time of the pulse whose pulse height is going to be estimated
double tstartnext
Starting time of the next pulse whose pulse height is going to be estimated
int lastPulse
It is 1 if the pulse is the last one into the record or the only one
double lrs
Running sum length (equal to the
LrsTinput parameter in samples)double lb
Baseline averaging length used for the pulse whose pulse height is going to be estimated
double B
In general, sum of the Lb digitized data samples (
LbTinput parameters in samples) of a pulse-free interval immediately before the current pulseint sizepulse
Size of the pulse in samples
double* pulseheight
Estimated pulse height of the pulse
-
int getSamplingrate_trigreclength_Filei(char *inputFile, struct Parameters par, double *samplingrate, int *trigreclength)¶
Located in file: initSIRENA.c
This function gets the sampling rate and the trig_reclength from an inputs FITS file
inputFile.Steps:
Open FITS file
Check if input FITS file have been simulated with TESSIM or XIFUSIM
Check if input XML file and XMl file to build the library to be used to reconstruct are the same
Get the sampling rate from the HISTORY keyword from the input FITS file and check with sampling rate from XML file
If xifusim file => Get ‘trig_reclength’ from the HISTORY keyword from the input FITS file ‘trig_reclength’ is necessary if SIRENA is going to run in THREADING mode
Close FITS file
Free memory
Members/Variables
char* inputFile
Input file name
struct Parameters par
Input parameters
double* samplingrate
(In) Sampling rate from XML file => (Out) Sampling rate
int* trigreclength
Necessary if SIRENA is going to run in THREADING mode
-
int getSamplingrate_trigreclength(char *inputFile, struct Parameters par, double *samplingrate, int *trigreclength, int *numfits)¶
Located in file: initSIRENA.c
This function gets the sampling rate and the trig_reclength no matter if
inputFileis only a FITS file or more (inputFile can start with ‘@’ or not, input file or files can have been simulated with TESSIM or XIFUSIM).Members/Variables
char* inputFile
Input file name
struct Parameters par
Input parameters
double* samplingrate
(In) Sampling rate from XML file => (Out) Sampling rate
int* trigreclength
Necessary if SIRENA is going to run in THREADING mode
int* numfits
Number of FITS files to work with
-
void gsl_vector_complex_absIFCA(gsl_vector *cvnew, gsl_vector_complex *cv)¶
Located in file: genutils.cpp
This function calculates the magnitude of the complex elements of a vector (real part).
Members/Variables
gsl_vector_complex* cv
Input GSL complex vector
gsl_vector* cvnew
Output GSL vector with the absolute values of the elements of
cv
-
void gsl_vector_complex_argIFCA(gsl_vector *varg, gsl_vector_complex *vin)¶
Located in file: genutils.cpp
This function calculates the arguments of the complex elements of a vector.
Members/Variables
gsl_vector_complex* vin
Input GSL complex vector
gsl_vector* varg
Output GSL vector with the arguments of the elements of
vin
-
void gsl_vector_complex_scaleIFCA(gsl_vector_complex *cv, gsl_complex z)¶
Located in file: genutils.cpp
This function multiplies the complex elements of a vector by a complex number.
Members/Variables
gsl_vector_complex* cv
Input/Output (scaled) GSL complex vector
gsl_complex z
Input GSL complex number
-
void gsl_vector_sqrtIFCA(gsl_vector *cvnew, gsl_vector *cv)¶
Located in file: genutils.cpp
This function calculates the square root of the elements of a vector.
Members/Variables
gsl_vector* cv
Input GSL complex vector
gsl_vector* cvnew
Output GSL vector with the square root values of the elements of
cv
-
int gsl_vector_Sumsubvector(gsl_vector *invector, long offset, long n, double *sum)¶
Located in file: genutils.cpp
This function returns the sum of some elements of the input vector.
The starting element of the sum is
offsetfrom the start of the input vector. It will sum upnelements.offsetcan take values from 0 to invector->sizeMembers/Variables
gsl_vector* invector
Input GSL vector
long offset
It is the first element to be summed
long n
Number of elements in the sum
double* sum
Calculated output value (sum of the corresponding elements)
-
int inDataIterator(long totalrows, long offset, long firstrow, long nrows, int ncols, iteratorCol *cols, void *user_strct)¶
Located in file: gennoisespec.cpp
This function takes the optimum number of rows to read the input FITS file and works iteratively
Steps:
Declare variables
Allocate input GSL vectors
Read iterator
- Processing each record
Information has been read by blocks (with nrows per block)
Just in case the last record has been filled out with 0’s => Last record discarded
Convert to the resistance space if necessary
To avoid taking into account the pulse tails at the beginning of a record as part of a pulse-free interval
Low-pass filtering
Differentiate
Finding the pulses: Pulses tstarts are found (call
findPulsesNoise())- Finding the pulse-free intervals in each record
If there are pulses => Call
findInterval()No pulses => The whole event is going to be used (DIVIDING into intervals of intervalMinBins size) => Call
findIntervalN()
Calculating the mean and sigma of the intervals without pulses together => BSLN0 and NOISESTD
Preparing the CSD calculus (not filtered data)
Free allocated GSL vectors
Members/Variables
long totalrows
Total number of rows processed
long offset
If positive, this number of rows at the beginning of the table (or pixels in the image) will be skipped and will not be passed to the work function
long firstrow
First row to read
long nrows
It specifies the number of table rows that are to be passed to the work function on each iteration. If nrows = 0 then CFITSIO will calculate the optimum number for greatest efficiency. If nrows is negative, then all the rows or pixels will be passed at one time, and the work function will only be called once. If any variable length arrays are being processed, then the nrows value is ignored, and the iterator will always process one row of the table at a time
int ncols
Number of columns
iteratorCol* cols
Structure of iteration
void* user_strct
This is a user supplied pointer that can be used to pass ancillary information from the driver routine to the work function. It may point to a single number, an array, or to a structure containing an arbitrary set of parameters
-
extern_C_void initializeReconstructionSIRENA(ReconstructInitSIRENA *reconstruct_init, char *const record_file, fitsfile *fptr, char *const library_file, char *const event_file, int pulse_length, double scaleFactor, int samplesUp, int samplesDown, double nSgms, int detectSP, int opmode, char *detectionMode, double LrsT, double LbT, char *const noise_file, char *filter_domain, char *filter_method, char *energy_method, double filtEev, double Ifit, char *ofnoise, int lagsornot, int nLags, int Fitting35, int ofiter, char oflib, char *ofinterp, char *oflength_strategy, int oflength, char preBuffer, double monoenergy, char hduPRECALWN, char hduPRCLOFWM, int largeFilter, int interm, char *const detectFile, int errorT, int Sum0Filt, char clobber, int maxPulsesPerRecord, double SaturationValue, char *const tstartPulse1, int tstartPulse2, int tstartPulse3, double energyPCA1, double energyPCA2, char *const XMLFile, int *const status)¶
Located in file: integraSIRENA.cpp
This function initializes the structure ReconstructInitSIRENA with the variables required for SIRENA reconstruction. The values are taken from the input parameters.
Load LibraryCollection structure if library file exists
Load NoiseSpec structure
Fill in the matrix tstartPulse1_i if tstartPulse1 = nameFile Start time (in samples) of the first pulse (0 if detection should be performed by the system; greater than 0 if provided by the user) or file name containing the tstart (in seconds) of every pulse,
tstartPulse1Fill in reconstruct_init
Members/Variables
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
char* const record_file
Filename of input data file with records,
RecordFilefitsfile* fptr
FITS object with pointer to data file
char* const library_file
File name of calibration library,
LibraryFilechar* const event_file
File name of output events (with reconstructed energy),
TesEventFileint pulse_length
Pulse length,
OFLengthNotPaddeddouble scaleFactor
Detection scale factor for initial filtering,
scaleFactorint samplesUp
Number of samples for threshold trespassing,
samplesUpint samplesDown
Number of samples below the threshold to look for other pulse,
samplesDowndouble nSgms
Number of standard deviations in the kappa-clipping process for threshold estimation,
nSgmsint detectSP
Detect secondary pulses (1) or not (0),
detectSPint opmode
Calibration run (0) or energy reconstruction run (1),
opmodechar* detectionMode
Adjusted Derivative (AD) or Single Threshold Crossing (STC),
detectionModedouble LrsT
Running sum length for the RS raw energy estimation (seconds),
LrsTdouble LbT
Baseline averaging length (seconds),
LbTchar* const noise_file
Noise file,
NoiseFilechar* filter_domain
Filtering Domain: Time (T) or Frequency (F),
FilterDomainchar* filter_method
Filtering Method: F0 (deleting the zero frequency bin) or F0 (deleting the baseline),
FilterMethodchar* energy_method
Energy calculation Method: OPTFILT, WEIGHT, WEIGHTN, I2R, I2RFITTED or PCA,
EnergyMethoddouble filtEev
Energy of the filters of the library to be used to calculate energy (only for OPTFILT, I2R and I2RFITTED),
filtEeVdouble Ifit
Constant to apply the I2RFITTED conversion
char* ofnoise
For optimal filtering: NSD or WEIGHTM,
OFNoiseint lagsornot
Lags (1) or no lags (0),
LagsOrNotint nLags
Number of lags (positive odd number)
int Fitting35
Number of lags to analytically calculate a parabola (3) or to fit a parabola (5)
int ofiter
Iterate (1) or not iterate (0),
OFIterchar oflib
Work or not with a library with optimal filters (1/0)
char* ofinterp
Optimal Filter by using the Matched Filter or the DAB as matched filter (MF/DAB) It has been fixed in tesreconstruction as DAB
char* oflength_strategy
Optimal Filter length Strategy: FREE, BYGRADE or FIXED,
OFStrategyint oflength
Optimal Filter length (taken into account if
OFStrategy= FIXED),OFLengthchar preBuffer
Some samples added or not before the starting time of a pulse (number of added samples read from the xml file)
double monoenergy
Monochromatic energy of input file in eV (only for library creation,
opmode= 0),monoenergyint hduPRECALWN
Add or not the PRECALWN HDU in the library file (yes/no) (only for library creation,
opmode= 0),hduPRECALWNint hduPRCLOFWM
Add or not the PRCLOFWM HDU in the library file (yes/no) (only for library creation,
opmode= 0),hduPRCLOFWMint largeFilter
Length of the longest fixed filters (only for library creation,
opmode= 0),largeFilterint interm
Write or not intermediate files (1/0),
intermediatechar* const detectFile
Intermediate detections file (if
intermediate= 1),detectFileint errorT
Additional error (in samples) added to the detected time (logically, it changes the reconstructed energies)
int Sum0Filt
0-padding: Subtract the sum of the filter (1) or not (0)
char clobber
Overwrite or not output files if exist (yes/no),
clobberint maxPulsesPerRecord
Default size of the event list,
EventListSizedouble SaturationValue
Saturation level of the ADC curves
int tstartPulse1
Start time (in samples) of the first pulse (0 if detection should be performed by the system; greater than 0 if provided by the user) or file name containing the tstart (in seconds) of every pulse,
tstartPulse1int tstartPulse2
Tstart (samples) of the second pulse,
tstartPulse2int tstartPulse3
Tstart (samples) of the third pulse (if 0 => PAIRS, if not 0 => TRIOS),
tstartPulse3double energyPCA1
First energy (only for
EnergyMethod= PCA)double energyPCA2
Second energy (only for
EnergyMethod= PCA)char * const XMLFile
File name of the XML input file with instrument definition
int* const status
Input/Output status
-
int InitialTriggering(gsl_vector *derivative, double nSgms, double scalefactor, double samplingRate, double stopcriteriamkc, double kappamkc, bool *triggerCondition, int *tstart, int *flagTruncated, double *threshold, int tstartProvided)¶
Located in file: pulseprocess.cpp
This function finds the first pulse in the input vector, first derivative of the (low-pass filtered) record.
Steps:
Declare variables
Stablish the
thresholdIt is necessary to find the tstart of the first pulse…
Obtain tstart of the first pulse in the derivative if
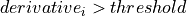
… Or to use the tstart provided as input parameter
Members/Variables
gsl_vector* derivative
First derivative of the (low-pass filtered) record
double nSgms
Number of Sigmas to establish the threshold (
nSgms)double scalefactor
Scale factor to calculate the LPF box-car length (
scaleFactor)double samplingRate
Sampling rate
double stopcriteriamkc
Used in
medianKappaClipping()(%)double kappamkc
Used in
medianKappaClipping()bool* triggerCondition
True => The algorithm has found the first event
False => The algorithm has not found any event
int* tstart
First event tstart (in samples)
int* flagTruncated
Flag indicating if the event is truncated
double* threshold
Calculated threshold (output parameter because it is necessary out of the function)
int tstartProvided
Tstart of the first pulse provided as input parameter
-
extern_C_void IntegrafreeTesEventListSIRENA(TesEventList *event_list)¶
Located in file integraSIRENA.cpp
This function frees the structure in the input parameter.
Members/Variables
TesEventList* event_list
Instance of TesEventList structure that contains the information of the reconstructed pulses
-
int interactivePars(inparam *taskPars, int np, string task)¶
Located in file inoututils.cpp
This function reads input parameters interactively (provided by the user or taken as default values). Used in tool gennoisespec.
Members/Variables
inparam* taskPars
Instance of inparam structure storing input parameters
int np
Number of parameters
string task
Tool name
-
int interpolatePOS(gsl_vector *x_in, gsl_vector *y_in, long size, double step, gsl_vector **x_out, gsl_vector **y_out)¶
Located in file: tasksSIRENA.cpp
This function interpolates an input vector (
x_in,y_in), creating an output vector (x_out,y_out) with the size and frequency step given. POS comes from the fact that the input spectrum only has positive frequencies (in order to not handle the f=0 bin).Declare and initialize variables
GSL method applied for interpolatation
Generate the interpolated output vector
Free memory
Members/Variables
gsl_vector* x_in
GSL input vector with the abscissas of the vector which is going to be interpolated
gsl_vector* y_in
GSL input vector with the ordinates of the vector which is going to be interpolated
long size
Size of the interpolated output vector
double step
Frequency step of the interpolated output vector
gsl_vector** x_out
GSL output vector with the abscissas of the interpolated vector
gsl_vector** y_out
GSL output vector with the ordinates of the interpolated vector
-
int interpolate_model(gsl_vector **modelFound, double p_model, gsl_vector *modelIn1, double p_modelIn1, gsl_vector *modelIn2, double p_modelIn2)¶
Located in file: pulseprocess.cpp
This function interpolates the pulse model,
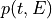 , between two models of the pulse models library,
, between two models of the pulse models library,
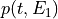 and
and 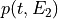 , being
, being 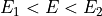 .
.According to the interpolation method:
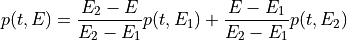
Members/Variables
gsl_vector** modelFound
Found model of the pulse whose energy or maxDER is
p_modeldouble p_model
Parameter (energy or maxDER) of the pulse whose model is being sought
gsl_vector* modelIn1
Model of the pulse whose parameter (energy or maxDER) is immediately lower than
p_modelin the library FITS filedouble p_modelIn1
Parameter (energy or maxDER) immediately lower than
p_modelin the library FITS filegsl_vector* modelIn2
Model of the pulse whose parameter (energy or maxDER) is immediately greater than
p_modelin the library FITS filedouble p_modelIn2
Parameter (energy or maxDER) immediately greater than
p_modelin the library FITS file
-
bool isNumber(string s)¶
Located in file: genutils.cpp
This function returns TRUE if the input string is a number or FALSE if not.
Members/Variables
string s
Input string
-
int loadRecord(TesRecord *record, double *time_record, gsl_vector **adc_double)¶
Located in file: tasksSIRENA.cpp
This fucntion loads the structure
recordinto theadc_doubleGSL vector.It checks if the record has been filled out with 0’s => It only loads the first values (which are different from 0).
Members/Variables
TesRecord* record
Member of TesRecord structure that contains the input record
double time_record
Starting time of the record (output)
gsl_vector** adc_double
Storage of the record to be processed (input/output)
-
int lpf_boxcar(gsl_vector **invector, int szVct, int sampleRate)¶
Located in file: pulseprocess.cpp
This function implements a low pass filtering as a box-car function in time.
The box-car function is a temporal average window:
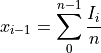
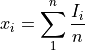
If the cut frequency of the filter is
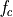 , the box-car length (n) is
, the box-car length (n) is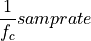
Steps:
Declare variables
Define the LPF (frequency domain) and the box-car function (time domain)
It is going to work with a longer vector to not have fake results for the last boxLength windows
Apply the box-car window by shifting it along the (lengthened) input vector
Free allocated GSL vectors
The function returns:
1: Function cannot run
3: Cut-off frequency too high => Equivalent to not filter
4: Cut-off frequency too low
Members/Variables
gsl_vector** invector
Input/Output GSL vector (non-filtered input vector/filtered input vector)
int szVct
Size of
invectorint sampleRate
Sampling rate (samples/s)
-
int matrix2vector(gsl_matrix *matrixin, gsl_vector **vectorout)¶
Located in file: tasksSIRENA.cpp
This function converts an input square matrix
![[n \times n]](_images/math/65c4484545603a3a8dfc82d595cc28b897032353.png) into an output
into an output 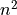 vector. It puts the first row of the matrix (
vector. It puts the first row of the matrix (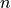 elements) in the first
elements) in the first 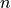 elements of the vector (from
elements of the vector (from  to
to 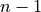 ), the second row of the matrix in the elements from
), the second row of the matrix in the elements from 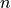 to
to 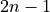 of the vector and so on.
of the vector and so on.Members/Variables
gsl_matrix* matrixin
GSL input square matrix
![[n \times n]](_images/math/65c4484545603a3a8dfc82d595cc28b897032353.png)
gsl_vector** vectorout
GSL output vector whose length is
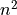
-
int medianKappaClipping(gsl_vector *invector, double kappa, double stopCriteria, double nSigmas, int boxLPF, double *threshold)¶
Located in file: pulseprocess.cpp
This function calculates a threshold in the first derivative of the record by using a Kappa-clipping method (replacing points beyond
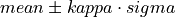 with the median).
with the median).Mean and sigma are calculated and values of
invectorout of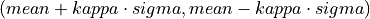 are replaced
with the median (it is trying to look for the baseline). And this process is iteratively repeated until there are
no points beyond
are replaced
with the median (it is trying to look for the baseline). And this process is iteratively repeated until there are
no points beyond 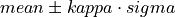 . Finally, the threshold is calculated as
. Finally, the threshold is calculated as 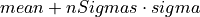 (‘+’ is used because
if there are pulses in the input invector they are always positive).
(‘+’ is used because
if there are pulses in the input invector they are always positive).Steps:
Declare variables
Calculate the median
Iterate until there are no points out of the maximum excursion (
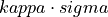 )
)Establish the threshold as mean+nSigmas*sigma
Members/Variables
gsl_vector* invector
First derivative of the (filtered) record
double kappa
Value to establish the range around of the mean
double stopCriteria
It is given in %
double nSigmas
Times sigma to calculate threshold as
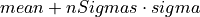
int boxLPF
Length of the low-pass filtering box-car
double* threshold
Calculated threshold
-
int medianKappaClipping_noiseSigma(gsl_vector *invector, double kappa, double stopCriteria, double nSigmas, double *mean, double *sigma)¶
Located in file: gennoisespec.cpp
This function provides the mean and the sigma of an input vector (with noise sigmas) by using a Kappa-clipping method (replacing points beyond
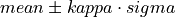 with the median).
with the median).First, mean and sigma are calculated and
invectorvalues out of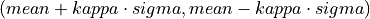 are replaced
with the median (it is trying to look for the baseline). And this process is iteratively repeated until there are
no points beyond
are replaced
with the median (it is trying to look for the baseline). And this process is iteratively repeated until there are
no points beyond 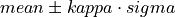 . Finally, the mean and sigma of the resulting vector are provided.
. Finally, the mean and sigma of the resulting vector are provided.Steps:
Declare variables
Calculate the median
Iterate until there are no points out of the maximum excursion (
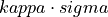 )
)Calculate mean and sigma
Members/Variables
gsl_vector* invector
First derivative of the (filtered) record
double kappa
Value to establish the range around of the mean
double stopCriteria
It is given in %
double nSigmas
Times sigma to calculate threshold as
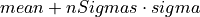
double* mean
Mean value of the
invector(no points beyond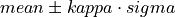 )
)double* sigma
Sigma value of the
invector(no points beyond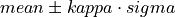 )
)
-
void MyAssert(int expr, char *msg)¶
Located in file: initSIRENA.c
This function displays an error message if the condition in
expris true.Members/Variables
int expr
Condition to be true in order to display the error message
char* msg
Message to be displayed
-
extern_C_OptimalFilterSIRENA *newOptimalFilterSIRENA(int *const status)¶
Located in file: integraSIRENA.cpp
Constructor. It returns a pointer to an empty OptimalFilterSIRENA data structure.
Members/Variables
int* const status
Input/output status
-
extern_C_PulsesCollection *newPulsesCollection(int *const status)¶
Located in file: integraSIRENA.cpp
Constructor. It returns a pointer to an empty PulsesCollection data structure.
Members/Variables
int* const status
Input/output status
-
extern_C_ReconstructInitSIRENA *newReconstructInitSIRENA(int *const status)¶
Located in file integraSIRENA.cpp
Constructor. It returns a pointer to an empty ReconstructInitSIRENA data structure.
Members/Variables
int* const status
Input/output status
-
int noDetect(gsl_vector *der, ReconstructInitSIRENA *reconstruct_init, int *numberPulses, gsl_vector **tstartgsl, gsl_vector **flagTruncated, gsl_vector **maxDERgsl, gsl_vector **samp1DERgsl)¶
Located in file: pulseprocess.cpp
This function runs if the starting time of the pulses are agiven as input parameters (
tstartPulse1!= 0). It looks for the maximum of the derivative of the pulse and the average of the first 4 samples of the derivative of the pulse.Members/Variables
gsl_vector* der
First derivative of the (low-pass filtered) record
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
int* numberPulses
Number of events
gsl_vector** tstartgsl
Starting time of the events (in samples)
gsl_vector** flagTruncated
Flag indicating if the event is truncated (inside this function only initial truncated pulses are classified)
gsl_vector** maxDERgsl
Maximum of the derivative of the event
gsl_vector** samp1DERgsl
Average of the first 4 samples of the derivative of the event
-
int obtainRiseFallTimes(gsl_vector *recordNOTFILTERED, double samprate, gsl_vector *tstartgsl, gsl_vector *tendgsl, gsl_vector *Bgsl, gsl_vector *Lbgsl, int numPulses, gsl_vector **tauRisegsl, gsl_vector **tauFallgsl)¶
Located in file: tasksSIRENA.cpp
This function provides an estimation of the rise and fall time of the detected pulses in a record.
Find the maximum of each pulse: amax
Baseline of each pulse: abase
- Find the first sample in the rising part above the 10% and 50%: t10 and t50
Line by using 2 points: (t10,a10) and (t50,a50)
t0 (t0,abase)
tmax (tmax,amax)
Rise time = tmax-t0
- Find the first sample in the decreasing part below the 50% and 10%: t50 and t10
Line by using 2 points: (t50,a50) and (t10,a10)
t0 (t0,abase)
tmax (tmax,amax)
Fall time = t0-tmax
Members/Variables
gsl_vector* recordNOTFILTERED
Record neither low-pass filtered nor differentiated
double samprate
Sampling rate
gsl_vector* tstartgsl
Starting time of the detected pulses in the record (samples)
gsl_vector* tendgsl
Ending time of the detected pulses in the record (samples)
gsl_vector* Bgsl
In general, sum of the Lb digitized data samples of a pulse-free interval immediately before each pulse
gsl_vector* Lbgsl
Number of samples added in Bgsl for each pulse
int numPulses
Number of detected pulses in the record
gsl_vector** tauRisegsl
Rise time of the detected pulses in the record (seconds)
gsl_vector** tauFallgsl
Fall time of the detected pulses in the record (seconds)
-
int parabola3Pts(gsl_vector *x, gsl_vector *y, double *a, double *b, double *c)¶
Located in file: genutils.cpp
This function calculates the equation of a parabola given 3 points.
Members/Variables
gsl_vector* x
Input GSL with x vector
gsl_vector* y
Input GSL with y vector
double* a
Fit coefficient of the quadratic term
double* b
Fit coefficient of the linear term
double* c
Fit coefficient (independent term)
-
int polyFit(gsl_vector *x_fit, gsl_vector *y_fit, double *a, double *b, double *c)¶
Located in file: genutils.cpp
This function makes a polynomial fitting
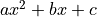 using the regression quadratic analysis. To measure how well model agrees with the data, the chi-square merit function is used, which in this case is
using the regression quadratic analysis. To measure how well model agrees with the data, the chi-square merit function is used, which in this case is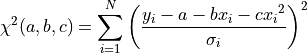
This equation is minimized to determine a, b and c. Then
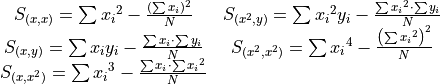
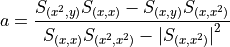
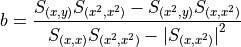
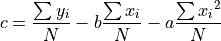
Members/Variables
gsl_vector* x_fit
Input GSL with x vector
gsl_vector* y_fit
Input GSL with y vector
double* a
Fit coefficient of the quadratic term
double* b
Fit coefficient of the linear term
double* c
Fit coefficient (independent term)
-
int polyFitLinear(gsl_vector *x_fit, gsl_vector *y_fit, double *a, double *b)¶
Located in file: genutils.cpp
This function makes a linear fitting
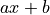 using the regression linear analysis. To measure how well model agrees with the data, the chi-square merit function is used, which in this case is
using the regression linear analysis. To measure how well model agrees with the data, the chi-square merit function is used, which in this case is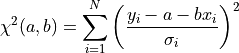
This equation is minimized to determine a and b. Then
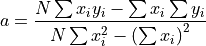
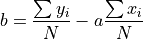
Members/Variables
gsl_vector* x_fit
Input GSL with x vector
gsl_vector* y_fit
Input GSL with y vector
double* a
Fit coefficient of the linear term
double* b
Fit coefficient (independent term)
-
void print_error(const char *const func, string message, int status)¶
Located in file: genutils.cpp
This function prints out error messages.
Members/Variables
const char* const func
Function name whose error is printed
string msg
Error message to be printed
int status
Status
-
int procRecord(ReconstructInitSIRENA **reconstruct_init, double tstartRecord, double samprate, fitsfile *dtcObject, gsl_vector *record, gsl_vector *recordWithoutConvert2R, PulsesCollection *foundPulses, long num_previousDetectedPulses, int pixid, gsl_vector *phid, int oscillations, int nrecord, double tstartPrevPulse)¶
Located in file: tasksSIRENA.cpp
This function processes the input record (detecting the pulses):
Declare and initialize variables
Allocate GSL vectors
(Low-pass filtering and) differentiation
If there are weird oscillations in the record, it is not processed => numPulses = 0
Find the events (pulses) in the record
If production mode (
opmode= 1):No detect if
tstartPulse1!= 0: ‘noDetect’Detect (
tstartPulse1!= 0):‘InitialTriggering’
‘FindSecondaries’ (
detectionMode= AD) or ‘FindSecondariesSTC’ (detectionMode= STC)
If calibration mode (
opmode= 0): ‘findPulsesCAL’
Calculate the end time of the found pulses and check if the pulse is saturated
Calculate the baseline (mean and standard deviation) before a pulse (in general before) => To be written in BSLN and RMSBSLN columns in the output FITS file
Obtain the approximate rise and fall times of each pulse
Load the found pulses data in the input/output foundPulses structure
Write test info (if reconstruct_init->intermediate = 1)
Write pulses info in intermediate output FITS file (if reconstruct_init->intermediate = 1)
Free allocated GSL vectors
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
double tstartRecord
Starting time of the record (in order to calculate absolute times)
double samprate
Sampling rate (in order to low-pass filter)
fitsfile* dtcObject
Object which contains information of the intermediate FITS file (to be written if
intermediate= 1)gsl_vector* record
GSL vector with signal values of input record
gsl_vector* recordWithoutConvert2R
GSL vector with original signal values of input record (without being converted to R space)
PulsesCollection* foundPulses
Input/output structure where the info about found pulses is stored
long num_previousDetectedPulses
Number of previous detected pulses (to know the index to get the proper element from tstartPulse1_i in case
tstartPulse1was a file name)int pixid
Pixel ID (from the input file) to be propagated
gsl_vector* phid
Photon ID (from the input file) to be propagated
int oscillations
1 (there are weird oscillations in the record) or 0 (record without weird oscillations)
int nrecord
Current record index
double tstartPrevPulse
tstart of the previous pulse (last pulse of the previous record) (seconds)
-
int pulseGrading(ReconstructInitSIRENA *reconstruct_init, int tstart, int grade1, int grade2, int *pulseGrade, long *OFlength, int nrecord)¶
Located in file: tasksSIRENA.cpp
This function provides the pulse grade (Pileup=-2, Rejected=-1, HighRes=1, MidRes=2, LimRes=3, LowRes=4) and the optimal filter length by taking into account the info read from the XML file and the
OFStrategy(FREE, BYGRADE or FIXED).Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
int tstart
Start time (samples)
int grade1
Pulse duration (length of optimal filter applied)
int grade2
Difference between the start time of the pulse and the start time of the previous pulse
int* pulseGrade
Pulse grade (output)
long* OFlength
Optimal filter length (=
OFLengthonly ifOFStrategy= FIXED andOFLength<= grade1) (output)
-
int readAddOrderParams(ReconstructInitSIRENA *reconstruct_init, fitsfile **inLibObject, double samprate, int eventcntLib, double estenergy, gsl_vector *pulsetemplate, gsl_matrix *covariance, gsl_matrix *weight, gsl_vector *pulsetemplateMaxLengthFixedFilter)¶
Located in file: tasksSIRENA.cpp
This function reads the library data, add new data (a new row) and sort the data according to an energy-ascending order.
Declare variables
Load values already in the library
Add new values
Realign
Add intermeadiate values
Recalculate intermediate values of some new pairs
Write values in the library
Free allocated GSL vectors
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
fitsfile** inLibObject
FITS object containing information of the library FITS file
double samprate
Sampling rate
int eventcntLib
Number of templates in the library
double estenergy
Pulse height of the template whose energy is going to be added to the library
gsl_vector* pulsetemplate
GSL vector with the pulse template whose energy is going to be added to the library
gsl_matrix* covariance
GSL matrix with covariance matrix of the energy which is going to be added to the library
gsl_matrix* weight
GSL matrix with weight matrix of the energy which is going to be added to the library
gsl_vector* pulsetemplateMaxLengthFixedFilter
GSL vector with the
largeFilter-length template whose energy is going to be added to the library
-
int readFitsComplex(IOData obj, gsl_matrix **result)¶
Located in file: inoututils.cpp
This function reads values of a complex column of a FITS file. After that, the function puts them into a GSL matrix for an easier processing.
Members/Variables
IOData obj
Input object for complex FITS column
gsl_matrix** result
Output GSL matrix
-
int readFitsSimple(IOData obj, gsl_vector **result)¶
Located in file: inoututils.cpp
This function reads values of a simple column of a FITS file. After that, the function puts them into a GSL vector for an easier processing.
Members/Variables
IOData obj
Input object for simple FITS column
gsl_vector** result
Output GSL vector
-
extern_C_void reconstructRecordSIRENA(TesRecord *record, TesEventList *event_list, ReconstructInitSIRENA *reconstruct_init, int lastRecord, int nRecord, PulsesCollection **pulsesAll, OptimalFilterSIRENA **optimalFilter, int *const status)¶
Located in file: integraSIRENA.cpp
This function is the main wrapper function to detect, grade and calculate the energy of the pulses in the input records.
Inititalize PulsesCollection structure
Check consistency of some input parameters
If first record, read the necessary keywords and columns from the input file in order to convert from current to quasi-resistance space
In case of running with threading
Detect pulses in input record (
runDetect()).- If reconstruction (
opmode= 1) and not PCA: Filter and calculate energy of pulses (
runEnergy())
- If reconstruction (
Fill in the
pulsesAllstructurePopulate output event list with pulses energies, arrival time and grading
Members/Variables
TesRecord* record
Instance of TesRecord structure that contains the input record
int trig_reclength
Record size (just in case threading and input files with different ADC lengths but the same record size indeed)
TesEventList* event_list
Instance of TesEventList structure that contains the information of the reconstructed pulses
ReconstructInitSIRENA* reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
int lastRecord
If record being analyzed is the last one,
lastRecord= 1. Otherwise it is equal to 0int nRecord
Input record number
PulsesCollection** pulsesAll
Member of PulsesCollection structure to successively store all the pulses used to create the library. Re-populated after each processed record.
OptimalFilterSIRENA** optimalFilter
Optimal filters used in reconstruction
int* const status
Input/output status
-
int RS_filter(gsl_vector *vector, double lrs, double lb, double B, double *pulseheight)¶
Located in file: pulseprocess.cpp
This function uses the running sum filter to find the pulse height. It always works in time domain.
A running sum filter, RS, is the sum of
lrsdigitized data samples. It is continuously updated upon the arrival of new data point. Simultaneously a baseline filter,B, is the sum oflbdigitized data samples without pulses. The algorithm looks for the time when RS/lrs reaches its maximum. At that time RS is stored,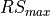 , and the baseline
is scaled with
, and the baseline
is scaled with lrs, Bp (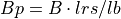 ). Then, the pulse height related to the pulse pseudoenergy is given by:
). Then, the pulse height related to the pulse pseudoenergy is given by: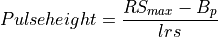
Members/Variables
gsl_vector* vector
Not filtered pulse (extracted from the record in
getPulseHeight())double lrs
Running sum length (samples)
double lb
Baseline averaging length (samples)
double B
In general, sum of the
lbdigitized data samples of a pulse-free interval immediately before the current pulsedouble* pulseheight
Pulseheight of the input pulse
-
void runDetect(TesRecord *record, int trig_reclength, int lastRecord, int nrecord, PulsesCollection *pulsesAll, ReconstructInitSIRENA **reconstruct_init, PulsesCollection **pulsesInRecord)¶
Located in file: tasksSIRENA.cpp
This function is responsible for the detection in SIRENA, record by record. It is used both for library creation (
opmode= 0) and energy reconstruction (opmode= 1) runnings.Conditions:
If first record and
opmode= 1 => RunfilderLibrary()If last record and
opmode= 0 => RuncalculateTemplate()andwriteLibrary()If
intermediate= 1 =>writeTestInfo()andwritePulses()If
opmode= 0 => Find pulses by usingfindPulsesCAL()If
opmode= 1 => Find pulses byInitialTriggering()andFindSecondaries()orFindSecondariesSTC()
Steps:
Create library file if it is necessary: calibration (
opmode= 0) and last record (runcreateLibrary())Create intermediate output FITS file if required (
createDetectFile())(Filter and) differentiate the models of the library (only for the first record in
opmode= 1). Run (filderLibrary())Store the input record in invector (
loadRecord())Detect weird oscillations in some GSFC records providing a warning (no pulses detected in that record)
Convert I into R if
EnergyMethod= I2R or I2RFITTED (convertI2R())Process each record (
proceRecord()):(Low-pass filter and) differentiate
Find pulses
Load the found pulses data in the input/output foundPulses structure
Write test info in intermediate output FITS file if
intermediate= 1 (writeTestInfo())Write pulses info in intermediate output FITS file if
intermediate= 1 (writePulses())
From this point forward, I2R and I2RFITTED are completely equivalent to OPTFILT
If last record in
opmode= 0 run:If last record and PCA:
In order to not have restrictions when providing (*reconstruct_init)->energyPCAx
Covariance data
Eigenvalues and eigenvectors
RSxN (S=2)
AE straight line: Pto0(x,y) and Pto10(x,y)
Calculus of the rotation angle
Rotation
Histograms of the two clusters (two energies)
Conversion factor from arbitrary unit to eV
Energy calculation
Close intermediate output FITS file if it is necessary
Members/Variables
TesRecord* record
Member of TesRecord structure that contains the input record
int trig_reclength
Record size (just in case threading and input files with different ADC lengths but the same record size indeed)
int lastRecord
Integer to verify whether record is the last one (=1) to be read (and thus if library file will be created)
PulsesCollection* pulsesAll
Member of PulsesCollection structure to successively store all the pulses used to create the library. Re-populated after each processed record
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
PulsesCollection** pulsesInRecord
Member of PulsesCollection structure to store all the pulses found in the input record
-
void runEnergy(TesRecord *record, int lastRecord, int nrecord, int trig_reclength, ReconstructInitSIRENA **reconstruct_init, PulsesCollection **pulsesInRecord, OptimalFilterSIRENA **optimalFilter)¶
Located in file: tasksSIRENA.cpp
This function calculates the pulse energy applying different methods (from
EnergyMethodandOFNoise). It only runs in RECONSTRUCTION mode (opmode= 1) (except toEnergyMethod= PCA).Declare variables
Store the
recordin invector (loadRecord())Subtract the baseline if
EnergyMethod= OPTFILT and runF0orB0val = 1 (FilterMethod= B0)Subtract the baseline if
EnergyMethod= WEIGHTCheck Quality
For each pulse:
Establish the pulse grade (for example VeryHighRes=1, HighRes=2, IntRes=3, MedRes=4, LimRes=5, LowRes=6, Rejected=-1, Pileup=-2) and the optimal filter length
Pulse: Load the proper piece of the record in pulse
- Get the low resolution energy estimator by filtering with a 8-samples-length (with lags) filter:
Load the low resolution pulse in pulse_lowres
Get the filter
Calculate the low resolution estimator
If
OFIter= 1, in the first iteration ( numiteration = 0) the values of maxDER and maxDERs are used infind_matchedfilterDAB(),find_optimalfilterDAB()orfind_Esboundary()getting the values of the energies which straddle the maxDER (Ealpha and Ebeta). It will have more iterations if the calculated energy is out of [Ealpha, Ebeta]. If energy is in [Ealpha, Ebeta] the iterative process stops.If
EnergyMethod= OPTFILT (or I2R, I2RFITTED) and reconstruct_init->OFLib = 0 andOFNoise= NSD:Find the matched filter and load it in filter (
find_matchedfilterDAB())Calculate the optimal filter
If
EnergyMethod= OPTFILT (or I2R, I2RFITTED) and reconstruct_init->OFLib = 1 andOFNoise= NSD:If it is necessary, choose the base-2 system value closest (lower than or equal) to the pulse length
Find the optimal filter and load it in filter (
find_optimalfilterDAB())
If
EnergyMethod= WEIGHT or WEIGHTN:Get the indexes of the two energies which straddle the pulse (
find_Esboundary())If
EnergyMethod= WEIGHTN and reconstruct_init->OFLib = 1:Choose the base-2 system value closest (lower than or equal) to the pulse length
find_prclwn()to find the appropriate values of the PRECALWN HDU (PCLx columns)
If
EnergyMethod= OPTFILT (or I2R, I2RFITTED) and reconstruct_init->OFLib = 1 andOFNoise= WEIGHTM:Choose the base-2 system value closest (lower than or equal) to the pulse length
find_prclofwm()to find the appropriate values of the PRCLOFWM HDU (OFWx columns)
Subtract the sum of the filter if
EnergyMethod= OPTFILT,OFNoise= NSD,FilterDomain= T, 0-padding andSum0Filt=1Calculate the energy of each pulse
If using lags, it is necessary to modify the tstart of the pulse
In order to subtract the pulse model, it has to be located in the tstart with jitter and know its values in the digitized samples
Subtract the pulse model from the record
Write info of the pulse in the output intemediate file if
intermediate= 1
Free allocated GSL vectors
Members/Variables
TesRecord** record
Structure that contains the input ADC record
int lastRecord
If record being analyzed is the last one,
lastRecord= 1. Otherwise it is equal to 0int nRecord
Input record number
int trig_reclength
Record size (just in case threading and input files with different ADC lengths but the same record size indeed)
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
PulsesCollection* pulsesInRecord
Collection of pulses found in the current record
OptimalFilterSIRENA** optimalFilter
Optimal filters used in reconstruction
PulsesCollection* pulsesAll
Member of PulsesCollection structure to store all the pulses found in the input FITS file. To know the index to get the proper element from tstartPulse1_i in case
tstartPulse1was a file name
-
void th_runEnergy(TesRecord *record, int lastRecord, int nrecord, int trig_reclength, ReconstructInitSIRENA **reconstruct_init, PulsesCollection **pulsesInRecord, OptimalFilterSIRENA **optimalFilter)¶
Located in file: tasksSIRENA.cpp
This function is responsible for the reconstruction in SIRENA (instead of
runEnergy()) when the THREADING running option has been chosen (hardcoded at this moment).
-
int shiftm(gsl_vector *vectorin, gsl_vector *vectorout, int m)¶
Located in file: tasksSIRENA.cpp
This function returns as
vectoroutthevectorindelayedmsamples.Members/Variables
int m
Delay in samples
gsl_vector* vectorin
GSL vector with input vector
gsl_vector* vectorout
-
int shift_m(gsl_vector *vectorin, gsl_vector *vectorout, int m)¶
Located in file: tasksSIRENA.cpp
This function returns as
vectoroutthevectorinmoved forward bymsamples.Members/Variables
int m
Advance in samples
gsl_vector* vectorin
GSL vector with input vector
gsl_vector* vectorout
-
char *subString(const char *input, int offset, int len, char *dest)¶
Located in file: initSIRENA.cpp
This function extracts some elements from an array of characters.
Members/Variables
const char* input
Array of characters from which some elements are extracted
int offset
Offset
int len
Length (number of elements to extract)
char* dest
Array of characters into which the extracted characters are written
-
int teslib_main()¶
Located in file: teslib.c
This function is mainly a wrapper to pass a data file to the SIRENA tasks in order to build a library.
Steps:
Register HEATOOL
Reading all programm parameters by using PIL
Read XML info
getSamplingrate_trigreclength()=> Obtain the trig_reclength and the sampling rateSixt standard keywords structure
Open output FITS file
Initialize data structures
Read the grading data from the XML file and store it in reconstruct_init_sirena->grading
Build up TesEventList
Call SIRENA to build the library
Save GTI extension to event file
Free memory
The user must supply the following input parameters (tesreconstruction.par file).
Parameters:
char RecordFile
Record FITS file
If
RecordFilestarts with ‘@’ it provides a file text containing several record input FITS fileschar TesEventFile
Output event list file
char LibraryFile
File with calibration library
char NoiseFile
Noise FITS file with noise spectrum
char XMLFile
XML input FITS file with instrument definition
char preBuffer
Some samples added or not before the starting time of a pulse (number of added samples read from the XML file) SIRENA’s format XML file (grading=>pre,post and pB) or new format XML file (grading=>pre,post and filtlen)
- pre=494, post=8192, pB=1000 pre=494, post=7192, filtlen=8192
preBuffer=filtlen-post
int EventListSize
Default size of the event list
char clobber
Overwrite or not output files if exist (yes/no)
char history
Write program parameters into output file
double scaleFactor
Detection scale factor for initial filtering
int samplesUp
Number of consecutive samples up for threshold trespassing
double nSgms
Number of quiescent-signal standard deviations to establish the threshold through the kappa-clipping algorithm
double LrsT
Running sum length for the RS raw energy estimation (seconds)
double LbT
Baseline averaging length (seconds)
double monoenergy
Monochromatic energy of the pulses in the input FITS file in eV
char hduPRECALWN
Add or not the PRECALWN HDU in the library file (yes/no)
char hduPRCLOFWM
Add or not the PRCLOFWM HDU in the library file (yes/no)
int largeFilter
Length of the longest fixed filter
char FilterDomain
Filtering Domain: Time (T) or Frequency (F)
char FilterMethod
Filtering Method: F0 (deleting the zero frequency bin) or B0 (deleting the baseline)
char EnergyMethod
Energy calculation Method: OPTFILT, WEIGHT, WEIGHTN, I2R, I2RFITTED or PCA
double Ifit
Constant to apply the I2RFITTED conversion
int intermediate
Write or not intermediate files (1/0)
char detectFile
Intermediate detections file (if
intermediate= 1)
-
int tesrecons_main()¶
Located in file: tesrecons.c
This function is mainly a wrapper to pass a data file to the SIRENA tasks in order to reconstruct the energies.
Steps:
Register HEATOOL
Reading all programm parameters by using PIL
Read XML info
getSamplingrate_trigreclength()=> Obtain the trig_reclength and the sampling rateSixt standard keywords structure
Open output FITS file
Initialize data structures for pulse filtering
Read the grading data from the XML file and store it in reconstruct_init_sirena->grading
Build up TesEventList
Call SIRENA to build reconstruct the energies
Save GTI extension to event file
Free memory
The user must supply the following input parameters (tesreconstruction.par file).
Parameters:
char RecordFile
Record FITS file
If
RecordFilestarts with ‘@’ it provides a file text containing several record input FITS fileschar TesEventFile
Output event list file
char LibraryFile
File with calibration library
char XMLFile
XML input FITS file with instrument definition
char preBuffer
Some samples added or not before the starting time of a pulse (number of added samples read from the XML file) SIRENA’s format XML file (grading=>pre,post and pB) or new format XML file (grading=>pre,post and filtlen)
- pre=494, post=8192, pB=1000 pre=494, post=7192, filtlen=8192
preBuffer=filtlen-post
int EventListSize
Default size of the event list
char clobber
Overwrite or not output files if exist (yes/no)
char history
Write program parameters into output file
double scaleFactor
Detection scale factor for initial filtering
int samplesUp
Number of consecutive samples up for threshold trespassing
int samplesDown
Number of consecutive samples below the threshold to look for other pulse
double nSgms
Number of quiescent-signal standard deviations to establish the threshold through the kappa-clipping algorithm
char detectionMode
Adjusted Derivative (AD) or Single Threshold Crossing (STC)
int detectSP
Detect secondary pulses (1) or not (0)
double LbT
Baseline averaging length (seconds)
char FilterDomain
Filtering Domain: Time (T) or Frequency (F)
char FilterMethod
Filtering Method: F0 (deleting the zero frequency bin) or B0 (deleting the baseline)
char EnergyMethod
Energy calculation Method: OPTFILT, WEIGHT, WEIGHTN, I2R, I2RFITTED or PCA
double filtEeV
Energy of the filters of the library to be used to calculate energy (only for OPTFILT, I2R and I2RFITTED)
double Ifit
Constant to apply the I2RFITTED conversion
char OFNoise
Noise to use with Optimal Filtering: NSD or WEIGHTM
int LagsOrNot
Lags or no lags (1/0)
int nLags
Number of lags (positive odd number)
int Fitting35
Number of lags to analytically calculate a parabola (3) or to fit a parabola (5)
int OFIter
Iterate or not iterate (1/0)
int OFLib
Work or not with a library with optimal filters (yes/no)
char OFStrategy
Optimal Filter length Strategy: FREE, BYGRADE or FIXED
int OFLength
Optimal Filter length (taken into account if
OFStrategy= FIXED)int OFLengthNotPadded
Filter length not padded with 0s (only necessary when reconstructing with 0-padding)
int errorT
Additional error (in samples) added to the detected time (Logically, it changes the reconstructed energies )
int Sum0Filt
0-padding: Subtract the sum of the filter (1) or not (0)
char tstartPulse1
Integer number: Sample where the first pulse starts or nameFile: File where the tstart (seconds) of every pulse is
int tstartPulse2
Tstart (samples) of the second pulse
int tstartPulse3
Tstart (samples) of the third pulse (if 0 => PAIRS, if not 0 => TRIOS)
double energyPCA1
First energy (only for PCA)
double energyPCA2
Second energy (only for PCA)
int intermediate
Write or not intermediate files (1/0)
char detectFile
Intermediate detections file (if
intermediate= 1)
-
int tesreconstruction_main()¶
Located in file: tesreconstruction.c
This function is mainly a wrapper to pass a data file to the SIRENA tasks in order to build a library or reconstruct the energy of the incoming X-ray photons after their detection which remains here only to backwards compatibility.
Steps:
Register HEATOOL
Reading all programm parameters by using PIL
Read XML info
getSamplingrate_trigreclength()=> Obtain the trig_reclength and the sampling rateSixt standard keywords structure
Open output FITS file
Initialize data structures needed for pulse filtering
Read the grading data from the XML file and store it in reconstruct_init_sirena->grading
Build up TesEventList to recover the results of the reconstruction
Call SIRENA to build the library or reconstruct the energies
Save GTI extension to event file
Free memory
The user must supply the following input parameters (tesreconstruction.par file).
Parameters:
char RecordFile
Record FITS file
If SIRENA => If RecordFile starts with ‘@’ it provides a file text containing several record input FITS files
char TesEventFile
Output event list file
int OFLengthNotPadded
Filter length not padded with 0s (only necessary when reconstructing with 0-padding)
int EventListSize
Default size of the event list
char clobber
Overwrite or not output files if exist (yes/no)
char history
Write program parameters into output file
char LibraryFile
File with calibration library
double scaleFactor
Detection scale factor for initial filtering
int samplesUp
Number of consecutive samples up for threshold trespassing (only used in calibration run, and in production run with STC detection mode)
int samplesDown
Number of consecutive samples below the threshold to look for other pulse (only used in production run with STC detection mode)
double nSgms
Number of quiescent-signal standard deviations to establish the threshold through the kappa-clipping algorithm
int detectSP
Detect secondary pulses (1) or not (0)
double LrsT
Running sum length for the RS raw energy estimation (seconds) (only for library creation)
double LbT
Baseline averaging length (seconds)
double monoenergy
Monochromatic energy of the pulses in the input FITS file in eV (only for library creation)
char hduPRECALWN
Add or not the PRECALWN HDU in the library file (yes/no) (only for library creation)
char hduPRCLOFWM
Add or not the PRCLOFWM HDU in the library file (yes/no) (only for library creation)
int largeFilter
Length of the longest fixed filter (only for library creation)
int opmode
Calibration run (0) or energy reconstruction run (1)
char detectionMode
Adjusted Derivative (AD) or Single Threshold Crossing (STC)
char NoiseFile
Noise FITS file with noise spectrum
char FilterDomain
Filtering Domain: Time (T) or Frequency (F)
char FilterMethod
Filtering Method: F0 (deleting the zero frequency bin) or B0 (deleting the baseline)
char EnergyMethod
Energy calculation Method: OPTFILT, WEIGHT, WEIGHTN, I2R, I2RFITTED or PCA
double filtEeV
Energy of the filters of the library to be used to calculate energy (only for OPTFILT, I2R and I2RFITTED)
double Ifit
Constant to apply the I2RFITTED conversion
char OFNoise
Noise to use with Optimal Filtering: NSD or WEIGHTM
int LagsOrNot
Lags or no lags (1/0)
int nLags
Number of lags (positive odd number)
int Fitting35
Number of lags to analytically calculate a parabola (3) or to fit a parabola (5)
int OFIter
Iterate or not iterate (1/0)
int OFLib
Work or not with a library with optimal filters (yes/no)
char OFStrategy
Optimal Filter length Strategy: FREE, BYGRADE or FIXED
int OFLength
Optimal Filter length (taken into account if
OFStrategy= FIXED)char preBuffer
Some samples added or not before the starting time of a pulse (number of added samples read from the xml file)
int intermediate
Write or not intermediate files (1/0)
char detectFile
Intermediate detections file (if intermediate*=1)
int errorT
Additional error (in samples) added to the detected time (Logically, it changes the reconstructed energies )
int Sum0Filt
0-padding: Subtract the sum of the filter (1) or not (0)
char tstartPulse1
Integer number: Sample where the first pulse starts or nameFile: File where the tstart (seconds) of every pulse is
int tstartPulse2
Tstart (samples) of the second pulse
int tstartPulse3
Tstart (samples) of the third pulse (if 0 => PAIRS, if not 0 => TRIOS)
double energyPCA1
First energy (only for PCA)
double energyPCA2
Second energy (only for PCA)
char XMLFile
XML input FITS file with instrument definition
-
int tesreconstruction_main()
Located in file: tesreconstruction.c
This function is mainly a wrapper to pass a data file to the SIRENA tasks in order to build a library or reconstruct the energy of the incoming X-ray photons after their detection.
Steps:
Register HEATOOL
Reading all programm parameters by using PIL
Read XML info
getSamplingrate_trigreclength()=> Obtain the trig_reclength and the sampling rateSixt standard keywords structure
Open output FITS file
Initialize data structures needed for pulse filtering
Read the grading data from the XML file and store it in reconstruct_init_sirena->grading
Build up TesEventList to recover the results of the reconstruction
Call SIRENA to build the library or reconstruct the energies
Save GTI extension to event file
Free memory
The user must supply the following input parameters (tesreconstruction.par file).
Parameters:
char RecordFile
Record FITS file
If SIRENA => If RecordFile starts with ‘@’ it provides a file text containing several record input FITS files
char TesEventFile
Output event list file
int OFLengthNotPadded
Filter length not padded with 0s (only necessary when reconstructing with 0-padding)
int EventListSize
Default size of the event list
char clobber
Overwrite or not output files if exist (yes/no)
char history
Write program parameters into output file
char LibraryFile
File with calibration library
double scaleFactor
Detection scale factor for initial filtering
int samplesUp
Number of consecutive samples up for threshold trespassing (only used in calibration run, and in production run with STC detection mode)
int samplesDown
Number of consecutive samples below the threshold to look for other pulse (only used in production run with STC detection mode)
double nSgms
Number of quiescent-signal standard deviations to establish the threshold through the kappa-clipping algorithm
int detectSP
Detect secondary pulses (1) or not (0)
double LrsT
Running sum length for the RS raw energy estimation (seconds) (only for library creation)
double LbT
Baseline averaging length (seconds)
double monoenergy
Monochromatic energy of the pulses in the input FITS file in eV (only for library creation)
char hduPRECALWN
Add or not the PRECALWN HDU in the library file (yes/no) (only for library creation)
char hduPRCLOFWM
Add or not the PRCLOFWM HDU in the library file (yes/no) (only for library creation)
int largeFilter
Length of the longest fixed filter (only for library creation)
int opmode
Calibration run (0) or energy reconstruction run (1)
char detectionMode
Adjusted Derivative (AD) or Single Threshold Crossing (STC)
char NoiseFile
Noise FITS file with noise spectrum
char FilterDomain
Filtering Domain: Time (T) or Frequency (F)
char FilterMethod
Filtering Method: F0 (deleting the zero frequency bin) or B0 (deleting the baseline)
char EnergyMethod
Energy calculation Method: OPTFILT, WEIGHT, WEIGHTN, I2R, I2RFITTED or PCA
double filtEeV
Energy of the filters of the library to be used to calculate energy (only for OPTFILT, I2R and I2RFITTED)
double Ifit
Constant to apply the I2RFITTED conversion
char OFNoise
Noise to use with Optimal Filtering: NSD or WEIGHTM
int LagsOrNot
Lags or no lags (1/0)
int nLags
Number of lags (positive odd number)
int Fitting35
Number of lags to analytically calculate a parabola (3) or to fit a parabola (5)
int OFIter
Iterate or not iterate (1/0)
int OFLib
Work or not with a library with optimal filters (yes/no)
char OFStrategy
Optimal Filter length Strategy: FREE, BYGRADE or FIXED
int OFLength
Optimal Filter length (taken into account if
OFStrategy= FIXED)char preBuffer
Some samples added or not before the starting time of a pulse (number of added samples read from the xml file)
int intermediate
Write or not intermediate files (1/0)
char detectFile
Intermediate detections file (if intermediate*=1)
int errorT
Additional error (in samples) added to the detected time (Logically, it changes the reconstructed energies )
int Sum0Filt
0-padding: Subtract the sum of the filter (1) or not (0)
char tstartPulse1
Integer number: Sample where the first pulse starts or nameFile: File where the tstart (seconds) of every pulse is
int tstartPulse2
Tstart (samples) of the second pulse
int tstartPulse3
Tstart (samples) of the third pulse (if 0 => PAIRS, if not 0 => TRIOS)
double energyPCA1
First energy (only for PCA)
double energyPCA2
Second energy (only for PCA)
char XMLFile
XML input FITS file with instrument definition
-
int th_runDetect(TesRecord *record, int trig_reclength, int lastRecord, int nrecord, PulsesCollection *pulsesAll, ReconstructInitSIRENA **reconstruct_init, PulsesCollection **pulsesInRecord)¶
Located in file: tasksSIRENA.cpp
This function is responsible for the detection in SIRENA (instead of
runDetect()) when the THREADING running option has been chosen (hardcoded at this moment). It is used both for library creation (opmode= 0) and energy reconstruction (opmode= 1) runnings.
-
int toGslMatrix(void **buffer, gsl_matrix **matrix, long numCol, int numRow, int type, int eventini)¶
Located in file: inoututils.cpp
The function puts the values of the input buffer into an output GSL matrix. Columns and rows are input parameters.
Members/Variables
void** buffer
Input buffer with data
gsl_matrix** matrix
Output GSL matrix
long numCol
Number of columns
int numRow
Number of rows
int type
FITS type (TINT, TSHORT, TDOUBLE, etc.)
int eventini
Initial event to start writing
-
int toGslVector(void **buffer, gsl_vector **array, long nevent, int eventini, int type)¶
Located in file: inoututils.cpp
The function puts the values of the input buffer into an output GSL vector.
Members/Variables
void** buffer
Input buffer with data
gsl_vector** array
Output GSL vector
long nevent
Number of elements to store
int eventini
Initial element number
int type
FITS type (TINT, TSHORT, TDOUBLE, etc.)
-
int vector2matrix(gsl_vector *vectorin, gsl_matrix **matrixout)¶
Located in file: tasksSIRENA.cpp
This function converts an input
 vector into an output square matrix
vector into an output square matrix ![[n \times n]](_images/math/65c4484545603a3a8dfc82d595cc28b897032353.png) . It puts the first
. It puts the first  elements of the vector in the first row of the matrix, the second group of
elements of the vector in the first row of the matrix, the second group of  elements (from
elements (from 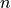 to
to 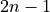 ) of the vector in the second row and so on.
) of the vector in the second row and so on.Members/Variables
gsl_vector** vectorin
GSL input vector whose length is
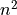
gsl_matrix* matrixout
GSL output square matrix
![[n \times n]](_images/math/65c4484545603a3a8dfc82d595cc28b897032353.png)
-
int weightMatrix(ReconstructInitSIRENA *reconstruct_init, bool saturatedPulses, PulsesCollection *pulsesAll, PulsesCollection *pulsesInRecord, long nonpileupPulses, gsl_vector *nonpileup, gsl_vector *pulseaverage, gsl_matrix **covariance, gsl_matrix **weight)¶
Located in file: tasksSIRENA.cpp
This function calculates the weight matrix by using the non piled-up pulses found in all the records, stored in pulsesAll (previous records) and pulsesInRecord (current record). The weight matrix of each energy (and other intermediate values) will be stored in the library by the function
fillInLibraryData().Definitions:
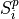 : Value of the ith-sample of the pulse number p
: Value of the ith-sample of the pulse number p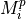 : Value of the ith-sample of the model number p (model= pulseaverage):
: Value of the ith-sample of the model number p (model= pulseaverage):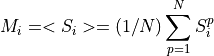
N: number of non piled-up pulses
![& D_i = S_i - M_i \\
& V_{ij} = <D_iD_j> = E[(S_i-M_i)(S_j-M_j)] = (1/N)\sum_{p=1}^{N}(S_i^p-M_i^p)(S_j^p-M_j^p) \\
& V = \left[\begin{matrix} <D_1D_1> & <D_1D_2> & ... & <D_1D_n> \\
<D_2D_1> & <D_2D_2> & ... & <D_2D_n> \\
.... & .... & ... & .... \\
<D_nD_1> & <D_nD_2> & ... & <D_nD_n>\end{matrix}\right]](_images/math/0630828451fea1a2f4657d36350e53d3effc83d8.png)
where n is the
OFLengthNotPaddedand thus![V = [n \times n]](_images/math/ef438dcdad8117f68c1f07ebaee17e6237c9cb25.png) .
.The weight matrix
![W = [V]^{-1}](_images/math/dadfd0c1c291e3beb691aaec067bfc0ae6915769.png) .
.Steps:
Calculate the elements of the diagonal of the covariance matrix
Calculate the elements out of the diagonal of the covariance matrix
If saturated pulses => Covariance matrix is a singular matrix => Non invertible
In order to allow the covariance matrix to be inverted => Replacing 0’s (0’s are due to the saturated values, equal in the pulse and in the model)
Elements of the diagonal: Generating a random double
 between a range (fMin,fMax) (-NoiseStd,NoiseStd) to replace 0’s with
between a range (fMin,fMax) (-NoiseStd,NoiseStd) to replace 0’s with 
Elements out of the diagonal: Generating two random doubles
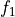 and
and 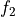 between a range (fMin,fMax) (-NoiseStd,NoiseStd) to replace 0’s with
between a range (fMin,fMax) (-NoiseStd,NoiseStd) to replace 0’s with 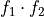
Calculate the weight matrix
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
bool saturatedPulses
If true, all the pulses (
opmode= 0 => all the pulses have the same energy) are saturatedPulsesCollection* pulsesAll
Collection of pulses found in the previous records
PulsesCollection* pulsesInRecord
Collection of pulses found in the current record
long nonpileupPulses
Number of non piled-up pulses
gsl_vector* nonpileup
GSL vector containing info about all the pulses informing if they are piled-up or not
gsl_vector** pulseaverage
GSL vector with the pulseaverage (= template = model) of the non piled-up pulses
gsl_matrix** covariance
GSL matrix with covariance matrix
gsl_matrix** weight
GSL matrix with weight matrix
-
int weightMatrixNoise(gsl_matrix *intervalMatrix, gsl_matrix **weight)¶
Located in file: gennoisespec.cpp
This function calculates the weight matrix of the noise
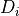 : Pulse free interval
: Pulse free interval
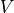 : Covariance matrix
: Covariance matrix![V_{ij} = E[DiDj]-E[Di]E[Dj]](_images/math/2b49d0667e1c339489b924c4bb35c2a1c8df7a86.png)
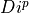 : Value of the pth-sample of the pulse-free interval i
: Value of the pth-sample of the pulse-free interval i
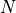 : Number of samples
: Number of samples![& V_{ij} = <D_iD_j> = E[D_iD_j] = (1/N)sum_{p=1}^{N}(Di^p)(Dj^p) \\
& V = \left[\begin{matrix} <D_1D_1> & <D_1D_2> & ... & <D_1D_n> \\
<D_2D_1> & <D_2D_2> & ... & <D_2D_n> \\
.... & .... & ... & .... \\
<D_nD_1> & <D_nD_2> & ... & <D_nD_n>\end{matrix}\right]](_images/math/1d218585bbb0f237c54f4118d633c19936d6aff3.png)
where n is the
OFLengthNotPaddedand thus![V = [n \times n]](_images/math/ef438dcdad8117f68c1f07ebaee17e6237c9cb25.png) .
.The weight matrix
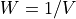 .
.Steps:
Calculate the elements of the diagonal of the covariance matrix
Calculate the elements out of the diagonal of the covariance matrix
Calculate the weight matrix
Members/Variables
gsl_matrix* intervalMatrix
GSL matrix containing pulse-free intervals whose baseline is 0 (baseline previously subtracted) [nintervals x intervalMinSamples]
gsl_matrix** weight
GSL matrix with weight matrix
-
int writeFilterHDU(ReconstructInitSIRENA **reconstruct_init, int pulse_index, double energy, gsl_vector *optimalfilter, fitsfile **dtcObject)¶
Located in file: tasksSIRENA.cpp
This function runs in RECONSTRUCTION mode and writes the optimal filter info (in the FILTER HDU) for each pulse if
intermediate= 1 and either reconstruct_init->OFLib = 0 or reconstruct_init->OFLib = 1,filtEeV= 0 and the the number of energies in the library FITS file is greater than 1.Declare variables
Open intermediate FITS file
- If (reconstruct_init->OFLib = 0) or (reconstruct_init->OFLib = 1,
filtEeV= 0 and the the number of energies in the library FITS file is greater than 1): Create the FILTER HDU if it is the first pulse
- Write data:
OPTIMALF or OPTIMALFF column (in time or frequency domain)
OFLENGTH column
- If (reconstruct_init->OFLib = 0) or (reconstruct_init->OFLib = 1,
Write ENERGY column in PULSES HDU
Close intermediate output FITS file if it is necessary
Free memory
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
int pulse_index
Index of the pulse whose info is going to be written (to know if it is the first pulse)
double energy
Estimated energy (eV)
gsl_vector* optimalfilter
Optimal filter (in time or frequency domain)
fitsfile** dtcObject
Fitsfile object for intermeadiate file name
-
int writeFitsComplex(IOData obj, gsl_matrix *matrix)¶
Located in file: inoututils.cpp
This function reads values of a GSL matrix. After that, the function puts them into a complex column of the output FITS file.
Members/Variables
IOData obj
Object for FITS column to be written
gsl_matrix* matrix
Input GSL matrix with data
-
int writeFitsSimple(IOData obj, gsl_vector *vector)¶
Located in file: inoututils.cpp
This function reads values of a GSL vector. After that, the function puts them into a column of the output FITS file.
Members/Variables
IOData obj
Object for FITS column to be written
gsl_vector* vector
Input GSL vector with data
-
int writeLibrary(ReconstructInitSIRENA *reconstruct_init, double samprate, double estenergy, gsl_vector *pulsetemplate, gsl_matrix *covariance, gsl_matrix *weight, bool appendToLibrary, fitsfile **inLibObject, gsl_vector *pulsetemplateMaxLengthFixedFilter)¶
Located in file: tasksSIRENA.cpp
This function writes the library (reordering if it is necesary and calculating some intermediate parameters)
Adding a new row to the library if appendToLibrary = true (
readAddSortParams())Write the first row of the library if appendToLibrary = false (
addFirstRow())In both cases, the keywords
CREADATEandSIRENAVwith the date and SIRENA version are written
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
double samprate
Sampling rate
double estenergy
Pulse height of the template whose energy is going to be added to the library
gsl_vector* pulsetemplate
GSL vector with the pulse template whose energy is going to be added to the library
gsl_matrix** covariance
GSL matrix with covariance matrix
gsl_matrix** weight
GSL matrix with weight matrix
bool appendToLibrary
true if adding a new row to the library and false if it is the first row to be added
fitsfile** inLibObject
FITS object containing information of the library FITS file
gsl_vector* pulsetemplateMaxLengthFixedFilter
GSL vector with the
largeFilter-length pulse template whose energy is going to be added to the library
-
void writeLog(FILE *fileRef, string type, int verbosity, string message)¶
Located in file: inoututils.cpp
This function includes the processing of the each level of message in the log file and the output screen:
Verbosity = 0 => The log file and the output screen include Errors
Verbosity = 1 => The log file and the output screen include Errors and Warnings
Verbosity = 2 => The log file and the output screen include Errors, Warnings and Alerts
Verbosity = 3 => The log file and the output screen include Errors, Warnings, Alerts and Log messages
Members/Variables
FILE* fileRef
File reference to log file
string type
String to indicate error type “Error”, “Warning”, “Alert”,”Log” or “OK”
int verbosity
Integer value for verbosity
string message
String message to print
-
int writePulses(ReconstructInitSIRENA **reconstruct_init, double samprate, double initialtime, gsl_vector *invectorNOTFIL, int numPulsesRecord, gsl_vector *tstart, gsl_vector *tend, gsl_vector *quality, gsl_vector *taurise, gsl_vector *taufall, fitsfile *dtcObject)¶
Located in file: tasksSIRENA.cpp
This function writes the data of the pulses found in the record in the intermediate FITS file (in the PULSES HDU). The pulses info given is: TSTART, I0 (the pulse itself), TEND, TAURISE, TAUFALL and QUALITY.
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values).
double samprate
Sampling rate (to convert samples to seconds)
double initialtime
Starting time of the record (in order to calculate absolute times)
gsl_vector* invectorNOTFIL
GSL vector with the original record (neither low-pass filtered nor differentiated)
int numPulsesRecord
Number of pulses found in the record
gsl_vector* tstart
GSL vector with the start times of the found pulses
gsl_vector* tend
GSL vector with the end times of the found pulses
gsl_vector* quality
GSL vector with the quality of the found pulses
0 => Standard (good) pulses
1 => Truncated pulses at the beginning
2 => Truncated pulses at the end
10 => Saturated pulses
11 => Truncated and saturated pulses
gsl_vector* taurise
GSL vector with the rise time constants of the found pulses (to be done)
gsl_vector* taufall
GSL vector with the fall time constants of the found pulses (to be done)
fitsfile* dtcObject
Object which contains information of the intermediate FITS file
-
int writeTestInfo(ReconstructInitSIRENA *reconstruct_init, gsl_vector *recordDERIVATIVE, double threshold, fitsfile *dtcObject)¶
Located in file: tasksSIRENA.cpp
This function writes the TESTINFO HDU in the intermediate FITS file. The written columns are FILDER (low-pass filtered and differentiated record) and THRESHOLD.
Members/Variables
ReconstructInitSIRENA** reconstruct_init
Member of ReconstructInitSIRENA structure to initialize the reconstruction parameters (pointer and values)
gsl_vector* recordDERIVATIVE
GSL vector with input record (low-pass filtered and) differentiated
double threshold
Threshold value used to find pulses
fitsfile dtcObject
Object which contains information of the intermediate FITS file
-
int writeTPSreprExten()¶
Located in file: gennoisespec.cpp
This function writes the noisespec output FITS file.
Steps:
Allocate GSL vectors
Write the data in the output FITS file (print only half of FFT to prevent aliasing)
NOISE HDU only contains positive frequencies (=> Multiply by 2 the amplitude)
NOISEALL HDU contains negative and positive frequencies => It is the HDU read to build the optimal filters
WEIGHTMS HDU
